- [2025-06-09] 🔥 stjudecab/rsvrecon v0.1 is released.
stjudecab/rsvrecon is a bioinformatics workflow developed to assemble and analyze genomic sequences of Respiratory Syncytial Virus (RSV) from Next-Generation Sequencing (NGS) data. It identifies genomic variations within RSV samples and highlights clinically relevant genomic features. To simplify interpretation, the workflow generates easy-to-understand HTML and PDF reports summarizing the results.
Built using Nextflow, the pipeline offers scalability, portability, and reproducibility
across diverse computational infrastructures. Dependency management is simplified by employing containerization
technologies such as Docker, Singularity, and Conda.
This pipeline utilizes the Nextflow DSL2 framework, featuring modularized processes with independent software environments, thereby making updates and maintenance straightforward. Processes are also integrated, whenever feasible, with the nf-core/modules repository to enhance usability and foster community contributions.
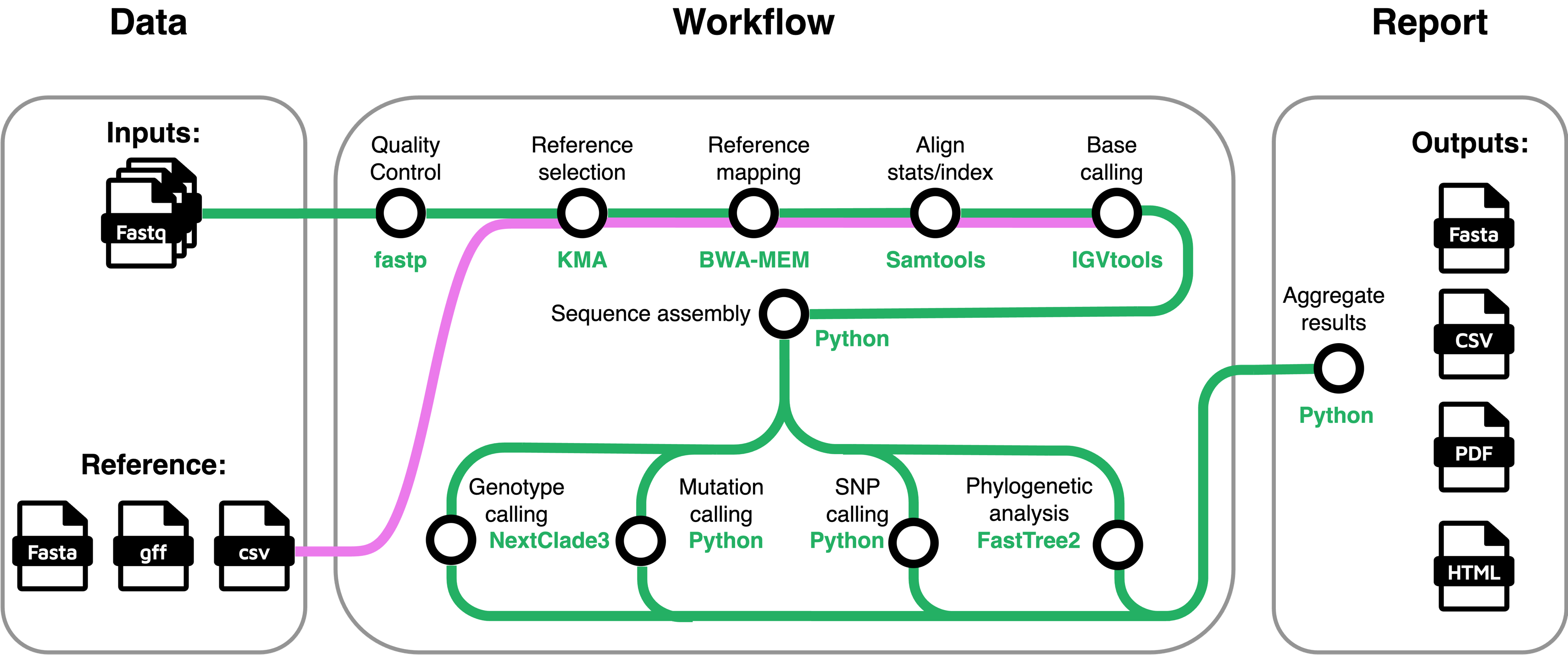
The schematic overview of the stjudecab/rsvrecon workflow is shown below:
Briefly, the rsvrecon pipeline performs the following major steps:
-
Merge Raw Reads (optional) Concatenate re-sequenced FastQ files (cat).
-
Quality Control of Raw Reads Evaluate sequencing quality (FastQC).
-
Adapter Trimming Remove adapters and low-quality bases (fastp).
-
RSV Database Mapping Map reads against RSV-specific databases (KMA).
-
Sequence Alignment
-
Genome Assembly
- Calculate genome coverage (IGVtools).
- Perform reference-guided genome assembly (custom script).
-
Variant Identification Determine viral clade assignments, mutations, and sequence quality (NextClade).
-
Genotyping (Whole Genome & G-gene)
Note
If you’re new to Nextflow or nf-core, please refer to the nf-core installation guide.
Run the workflow first with -profile test to ensure proper functionality before applying it to your actual data.
Prepare your sample metadata in a CSV format (samplesheet.csv) with FASTQ files:
sample,fastq_1,fastq_2
sample_1,sample_1_R1_001.fastq.gz,sample_1_R2_001.fastq.gz
sample_2,sample_2_R1_001.fastq.gz,sample_2_R2_001.fastq.gz
Note
If a sample has multiple sequencing lanes or replicates, list each replicate in a separate row with the
same sample ID. The pipeline will automatically merge these reads before analysis. Spaces in sample IDs will be
automatically converted to underscores (_).
Run the workflow using the following command structure:
# For St. Jude HPC users, specify the institutional profile, e.g., '-profile stjude'
nextflow run stjudecab/rsvrecon \
-profile <docker/singularity/.../institution_config> \
--input samplesheet.csv \
--outdir <OUTDIR> \
<args>Warning
Pipeline parameters should only be provided via CLI arguments or Nextflow's -params-file option. Custom
configuration files (-c) can specify system and pipeline configurations except for parameters.
For detailed guidance, consult the configuration documentation.
For further information, please check the complete usage documentation and pipeline output descriptions.
Pipeline results are organized per sample ID within the specified output directory (<OUTDIR>), structured by analysis stages:
<OUTDIR>/<sample_id>/fastq: Contains trimmed FASTQ files ready for alignment.<OUTDIR>/<sample_id>/bam: Includes aligned BAM files and index files (BAI).<OUTDIR>/<sample_id>/qc: Quality control outputs for raw and aligned reads.<OUTDIR>/<sample_id>/reference: Reference genomes and database files used for the analysis.<OUTDIR>/<sample_id>/variant_calling: Variant calling and identification results.<OUTDIR>/<sample_id>/phylogeny_tree: Results related to phylogenetic analyses.<OUTDIR>/<sample_id>/assembly: Genome assemblies and coverage summaries.- (and more)
In addition to sample-level results, our pipeline delivers batch-level results as well, structured as follows:
<OUTDIR>/batch_qcs/: Contains MultiQC-based QC reports for both raw and filtered (trimmed) data.<OUTDIR>/batch_reports/: Include combined results across samples as clinical reports in bothPDFandHTMLformats.
stjudecab/rsvrecon was developed by Haidong Yi (@HaidYi) and Lei Li (@LeiLi-Uchicago) at the Center for Applied Bioinformatics (CAB), St. Jude Children's Research Hospital. The pipeline design incorporates community-driven best practices, especially inspired by nf-core. We also thank the wider CAB team for their valuable inputs and feedbacks.
We encourage community contributions. Please adhere to nf-core guidelines for maintaining consistency. Suggestions and improvements are welcome through pull requests or issues on our GitHub repository.
The pipeline logo is initially generated through ChatGPT's
new 4o Image Generation function using the pipeline introduction as the prompt.
@article{Li2025.rsvrecon,
author = {Li, Lei and Yi, Haidong and Brazelton, Jessica N. and Webby, Richard and Hayden, Randall T. and Wu, Gang and Hijano, Diego R.},
title = {Bridging Genomics and Clinical Medicine: RSVrecon Enhances RSV Surveillance with Automated Genotyping and Clinically-important Mutation Reporting},
elocation-id = {2025.06.03.657184},
year = {2025},
doi = {10.1101/2025.06.03.657184},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2025/06/09/2025.06.03.657184},
journal = {bioRxiv}
}