Dirichlet Allocation of MUTAtions in cancer
Damage and Misrepair Signatures: Compact Representations of Pan-cancer Mutational Processes
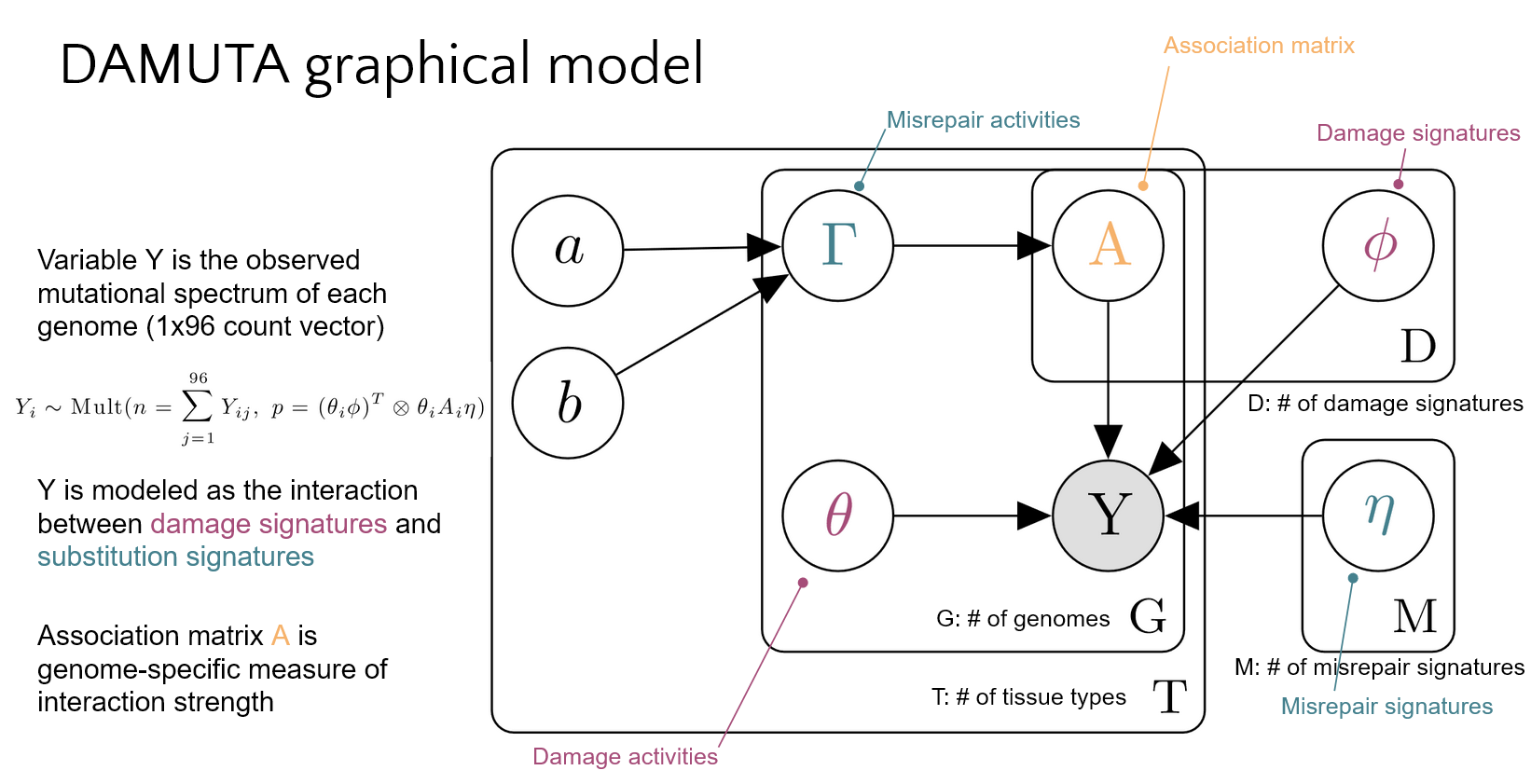
- Separately model damage and misrepair processes
- Estimate activities of DAMUTA signatures
- Fit new Damage- and Misrepair-signatures denovo
nb. internally these signatures are referred to by their symbols in the graphical model: eta and phi respectively.
- damuta python package
- docs package documentation
- example notebooks usage examples & tutorials
- manuscript code code to reproduce experiments and plots reported in manuscript
DAMUTA documentation is hosted via readthedocs
Recommended Requirements:
- Unix-based system (tested on CentOS Linux 7)
- RAM: 16+ GB (for large-scale pan-cancer datasets)
- CPU: 4+ cores, 3.0+ GHz/core
- GPU: NVIDIA GPU with CUDA support (optional, for GPU acceleration via Theano)
Software dependencies are specified in (damuta_env.yml)[./damuta_env.yml]
Clone this repo git clone https://github.com/morrislab/damuta
conda env create -f damuta_env.yml
conda activate damuta
pip install -e .pip install damutaTo use the GPU, ~/.theanorc should contain the following:
[global]
floatX = float64
device = cuda
Otherwise, device will default to CPU.
Conda environment setup: ~3-5 minutes Package installation: ~2-3 minutes
See quickstart to get started (~5 min)
See manuscript code for code to reproduce experiments and plots reported in manuscript
Some data files are omitted from this repository due to access restrictions. Access can be requested from the corresponding sources:
Unrestricted-access data and certain useful intemediate files are also available via can be downloaded from zenodo
To download and organize these data:
# in top-level directory
wget https://zenodo.org/records/15685052/files/damuta_zenodo.zip
unzip damuta_zenodo
mv damuta_zenodo/data/* manuscript/data
mv damuta_zenodo/figure_data/* manuscript/results/figure_data
# clean up now-empty directories
rmdir damuta_zenodo/data damuta_zenodo/figure_data damuta_zenodo
| file name | info | source |
|---|---|---|
| COSMIC_v3.2_SBS_GRCh37.csv | COSMIC database | |
| icgc_sample_annotations_summary_table.txt | sample annotations used by PCAWG heterogeneity & evolution working group | ICGC data portal |
| PCAWG_sigProfiler_SBS_signatures_in_samples | counts of mutations attributed to each signature for PCAWG samples | syn11738669.7 |
| pcawg_counts.csv | mutation type counts in PCAWG samples | Derived from syn7357330 |
| pcawg_cancer_types.csv | sample annotations used in Jiao et. al | Adapted from z-scores file |
| gel_clinical_ann.csv | tumour type annotations for 18640 samples (ICGC, HMF, GEL) | Adapted from Degasperi et. al table S6 |
| gel_counts.csv | mutation type counts for 18640 samples (ICGC, HMF, GEL) | Adapted from Degasperi et. al table S7 |
@misc{harrigan_damage_2025,
title = {Damage and {Misrepair} {Signatures}: {Compact} {Representations} of {Pan}-cancer {Mutational} {Processes}},
copyright = {© 2025, Posted by Cold Spring Harbor Laboratory. This pre-print is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), CC BY-NC 4.0, as described at http://creativecommons.org/licenses/by-nc/4.0/},
shorttitle = {Damage and {Misrepair} {Signatures}},
url = {https://www.biorxiv.org/content/10.1101/2025.05.29.656360v1},
doi = {10.1101/2025.05.29.656360},
language = {en},
urldate = {2025-06-02},
publisher = {bioRxiv},
author = {Harrigan, Caitlin F. and Campbell, Kieran and Morris, Quaid and Funnell, Tyler},
month = jun,
year = {2025},
}