Topic B : Detection of chromosome compartments
Description of the project: Project of the inter-university collaborative course Meet-EU 2021/2022. It concerns the detection of chromatin compartments, based on Hi-C intra chromosomal contacts maps in different resolutions. We present data pre-processing pipeline, already known and confirmed detection method of 2 basic chromosome compartments active (euchromatin) and inactive(heterochromatine) , as well as some methods to detect subcompartments and define their optimal number. For more information, please check: https://www.hdsu.org/meet-eu-2021/ https://www.cell.com/cell/fulltext/S0092-8674(14)01497-4
Installation and libraries requirements (Python 3):
HiCtoolbox.py (please use the version of HiCtoolbox uploaded here and not the original one from Léopold Carron repository, as we made some adjustments necessary so that our main code works correctly !) hmmlearn (https://hmmlearn.readthedocs.io/en/latest/api.html#hmmlearn-hmm) h5py (https://docs.h5py.org/en/stable/) sklearn (https://scikit-learn.org/stable/) scipy ( https://scipy.org/) standard Python libraries ( pandas, numpy, matplotlib,seaborn) We installed the specific tools via Anaconda (https://www.anaconda.com/products/individual) using conda install command in the Anaconda prompt.
USAGE:
Main code is in main.ipynb. It might take couple of minutes to run. Please pay attention to compartment marking in the exported file , in case you wish to make some comparisons. We mark 1 as an active compartement A, -1 as inactive compratement B , 0 as filtered bin . !
Datasets:
Hi-C contact map (we deliver: chr22_100kb.txt ) Gold-standard eigenvector compartments detection file ( we deliver: "chr22_VP_100kb.txt") Epigenetic marks file ("E116_15.bed") Gene density file ("chr22.hdf5" ) Please search for the datasets of your interest here: http://www.lcqb.upmc.fr/meetu/
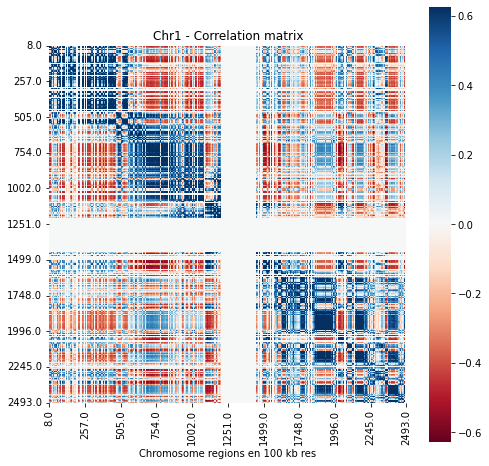
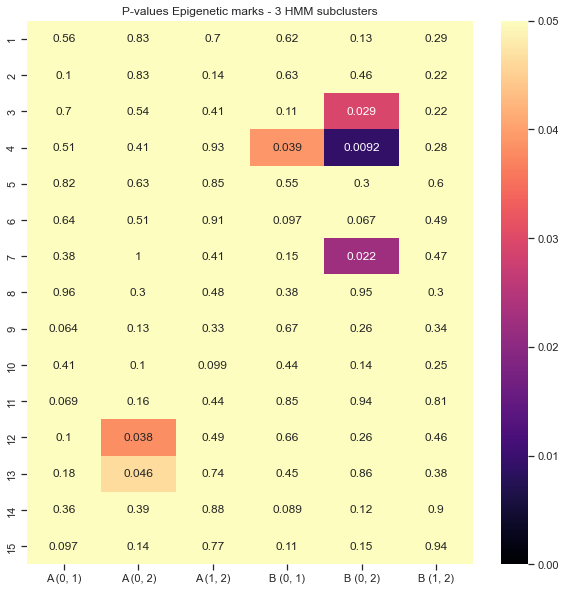
Results visualisation:
Authors and acknowledgments: Oktawia SCIBIOR, Mikal DAOU, Maxime GUEUDRE
Code credits to: Léopold Carron