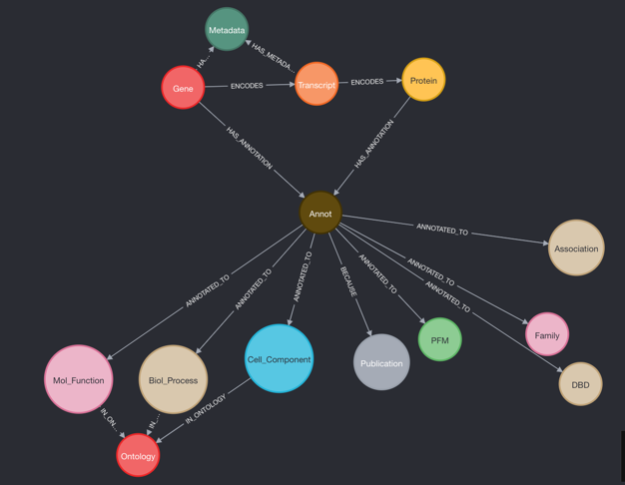
The graphTF project facilitates quick and comprehensive TF comparisons that consider multiple types/sources of data.
It does this by:
- Unifying Transcription Factor (TF) Annotations from high-value sources into a human and machine-readable Neo4j Graph Database
- Using the structure of the graph database to develop pairwise TF similarity scores. This provides an automated and quantitative measure of similarity between any 2 TFs.
Directories:
├── archived #old data files and scripts, not used in current graph model
│ ├── biogrid_example_scripts
│ └── exampleDataset
├── current #current working build of database
│ ├── construction #scripts for constructing and populating new neo4j database instance
│ │ ├── __pycache__
│ │ ├── config #neo4j database instance URI/Username/Passkey and links raw github datafiles
│ │ ├── core #Code to parse YML file in construction/config
│ │ │ └── __pycache__
│ │ └── cypher_scripts #scripting for reformatting raw data to graph schema
│ │ └── GO
│ ├── data #store of all data flowing in from outside databases and out to the graph database
│ │ ├── entities #gene, transcript, protein data
│ │ ├── gene_annotations #transcription factor annotation data
│ │ │ └── cis-bp
│ │ └── protein_interactions #protein interaction data
│ ├── doc #notes on learnings and descisions made during work on project
│ └── import #scripts for ingesting remote data into local csv files
│ ├── __pycache__
│ ├── config #URL and passkeys needed to interact w various biological databases
│ └── core #code to parse config YML file
│ └── __pycache__
└── imagesSchema:
Mapping genes to TF:
- Canonical Gene->Protein Relationships are chosen by canonical transcript status and primary sequence status in ensembl. These criteria yield a single gene->transcript->protein pathway for every protein in yamanaka graph.
Mapping annotations to TF:
- Protein associations and interaction attached to protein isoform.
- All other annotation attached to gene encoding TF.