Implementation of genomic selection in hybrid sweet corn breeding programs targeting long-term genetic gains
Marco Antônio Peixoto*1,2 , Kristen Leach 1 , Leonardo Bhering2 , Patrick Flannery3 , Jared Zystro4 , William Tracy3 , Márcio F. R. Resende1
Hybrid prediction through genomic selection (GS) is a tool that can reduce time and costs related with the generation of successful hybrids. The development of GS prediction models is expected to increase the probability of superior hybrids reaching advanced field-testing stages. In this study we explore the potential of implementing GS in a sweet corn breeding program through hybrid prediction in an across-year framework and we validate the implementation of genomic selection in a long-term in the sweet corn breeding program by stochastic simulations.
A total of 513 hybrids were created from a collaboration between the University of Florida and University of Wisconsin sweet corn breeding programs. Hybrids were assessed in three locations - Florida (FL), California (CA), and Wisconsin (WI) - in two consecutive years (2020-21). We compare the ability of two GS models (genomic best linear unbiased prediction -GBLUP- and reproducing kernel Hilbert space -RKHS) in single- and multi-trait framework (STM and MTM) for hybrid prediction. The models used accounted for additive (A) and additive + dominance (AD) effects. The traits assessed were as followed:
Table 1: Traits measured in each site and used in the model precition. √: trait measured.
| Traits | Short | Florida | California | Wisconsin |
|---|---|---|---|---|
| Ear length | EL | √ | √ | √ |
| Ear width | EW | √ | √ | √ |
| Tip fill | TPF | √ | √ | √ |
| Stand count | STAND | √ | √ | - |
| Days to pollination | DTP | - | √ | √ |
| Husk protection | HP | - | √ | √ |
| Kernel row number | KRN | - | - | √ |
| Plant height | PH | - | √ | - |
| Ear height | EH | - | √ | - |
| Solidity | SOL | √ | - | - |
| Taper | TP | √ | - | - |
| Curvature | CUR | √ | - | - |
| Days to silking | DTS | - | - | √ |
| Husk appearence | HAP | - | - | √ |
| Row appearence | RAP | - | - | √ |
| Ear shape | ES | - | - | √ |
| Color rate | CR | - | - | √ |
| Flavor | FL | - | - | √ |
| Texture | TXT | - | - | √ |
| Rating | RT | - | - | √ |
Stochastic simulation were used to compare the implementation of genomic selection into the sweet corn breeding program. We used the trait architecture information to perform a multi-trait breeding simulation for six traits that mimic important sweet corn fresh market traits: EH, LA, EL, EW, KRN, and GRM. Three scenarios where simulated:
- Conventional breeding program (CONV): A conventional sweet corn breeding program with phenotypic selection.
- Genomic selection breeding program (CONVGS): A conventional sweet corn breeding program with genomic selection.
- Double-haploid breeding program (DH): A sweet corn breeding program with double-haploid and genomic selection.
These scenarios were compared using the mean of 50 replicates, each replicate consisting of: i. a burn-in phase, used in all scenarios as common starting point with 20 years of breeding, and, ii. an advanced phase of evaluation comprising of 30 years.
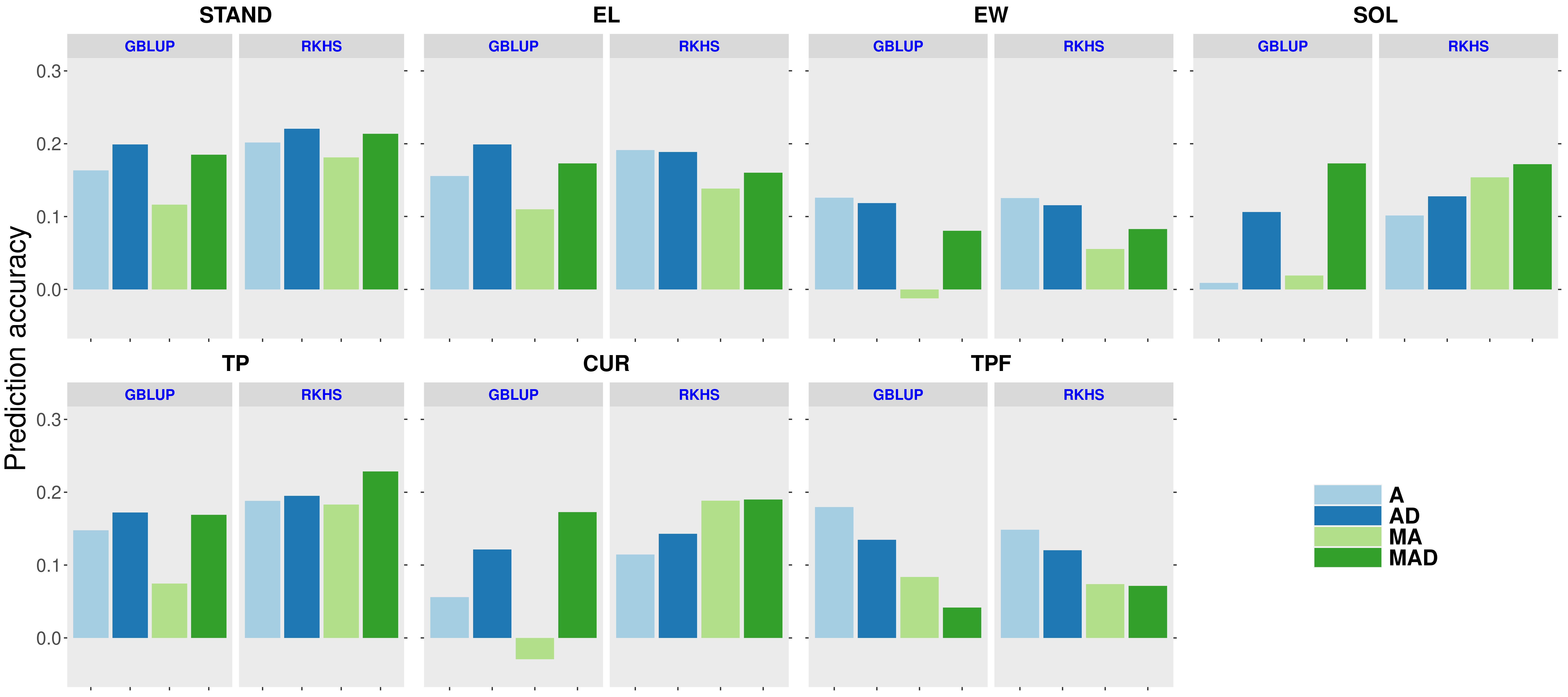
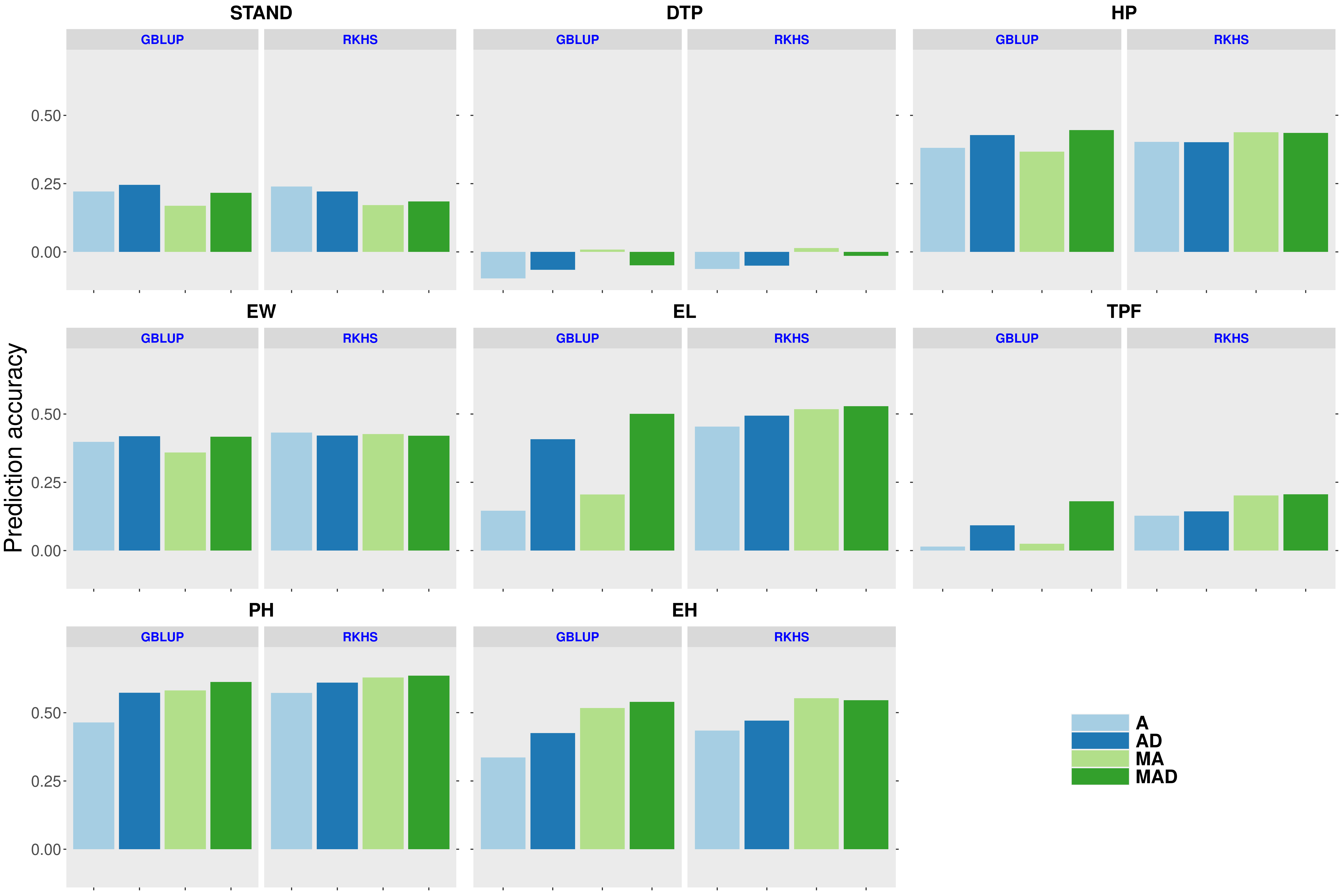
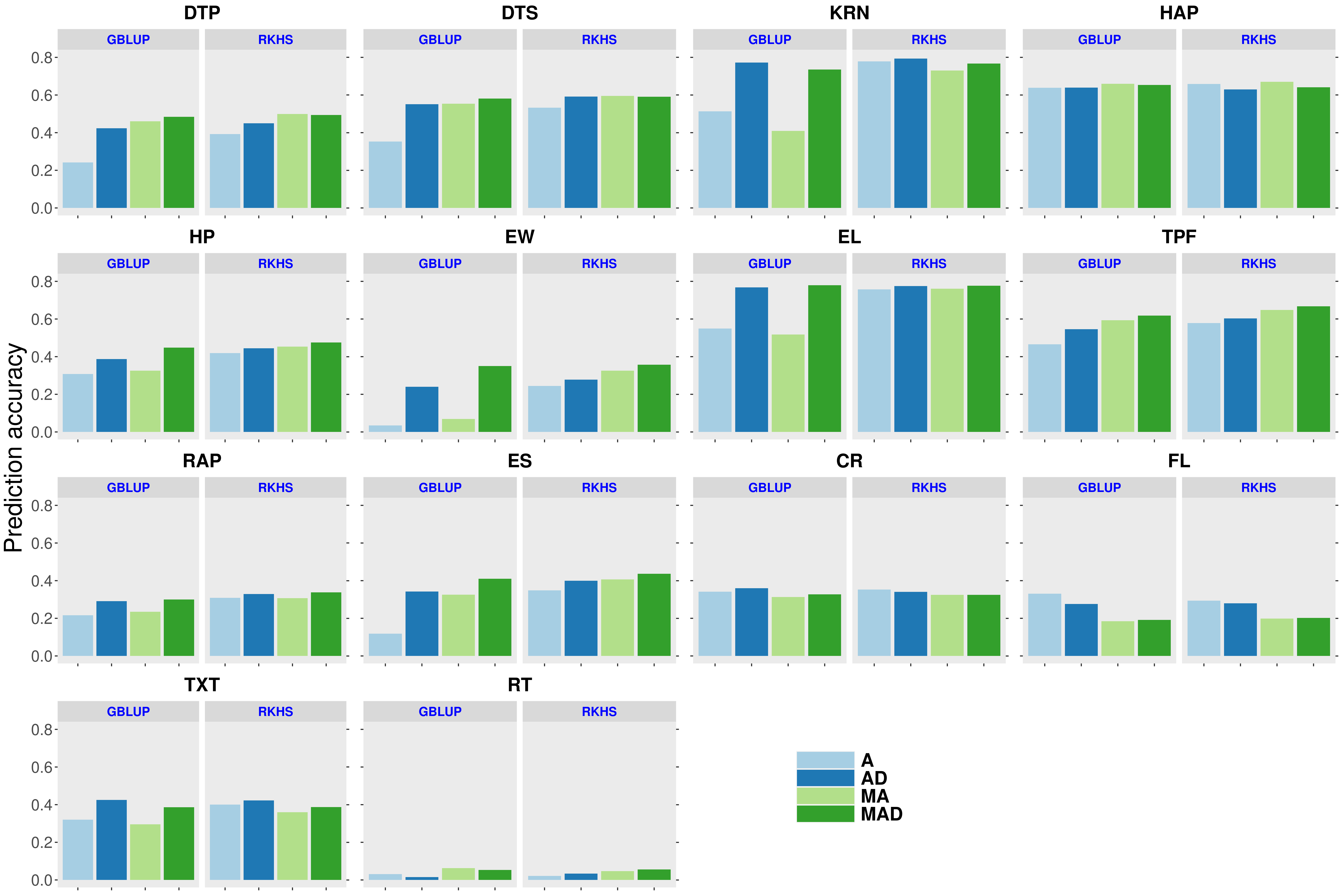
The RKHS models outperformed GBLUP models in across-year prediction (18%, 16%, 16% for CA, FL, and WI sites, respectively). The use of models accounting for AD effects increased the predictability in 15% (CA), 9% (FL), and 15% (WI) compared with the models with only A effects (Figures 1-3). Overall, the MTM outperformed STM by 11%, 15%, and 14%, for CA, FL, and WI, respectively.
Figure 1: In across-year hybrid prediction accuracy for the traits assessed in Florida site. A: single-trait additive model. AD: single-trait additive + dominance model. MA: multi-trait additive model. MAD: multi-trait additive + dominance model.
Figure 2: In across-year hybrid prediction accuracy for the traits assessed in California site. A: single-trait additive model. AD: single-trait additive + dominance model. MA: multi-trait additive model. MAD: multi-trait additive + dominance model.
Figure 3: In across-year hybrid prediction accuracy for the traits assessed in Wisconsin site. A: single-trait additive model. AD: single-trait additive + dominance model. MA: multi-trait additive model. MAD: multi-trait additive + dominance model.
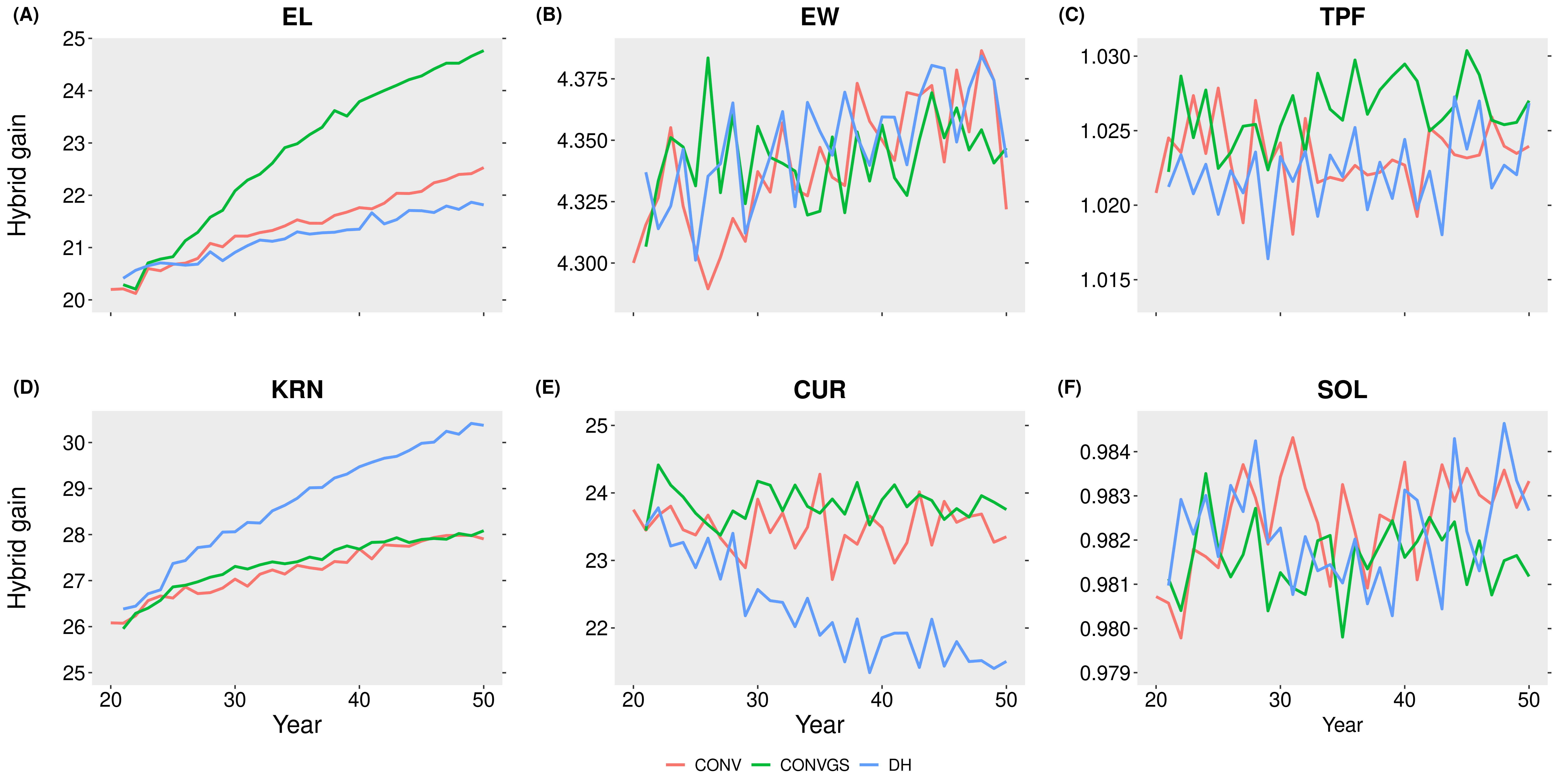
Simulations indicated that hybrid performance differed depending on the target trait considered. Generally, some traits presented large hybrid gains over time while other traits presented only small hybrid gains.
Figure 4. Hybrid gain overtime for the four simulated scenarios for the six traits simulated in a multitrait framework. The hybrid gain is plotted as a mean of the hybrids for each cycle. Each line represents the hybrid gain for the 50 replicates. CONV: A conventional sweet corn breeding program with phenotypic selection. CONVGS: A conventional sweet corn breeding program with genomic selection. DHGS: A sweet corn breeding program with double-haploid and genomic selection. EL: Ear length. EW: Ear width. TPF: tip fill. KRN: kernel row number. CUR: curvature. SOL: solidity.
This study show the potential of improving hybrid prediction in a sweet corn breeding program by considering RKHS, multitrait and, AD effects in a model. The application of genomic selection in a long-term framework returns large gains for some target traits.