Uniformizing Techniques to Process CT scans with 3D CNNs for Tuberculosis Prediction [arXiv]
This code is part of the supplementary materials for our paper which is published in the 23rd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI) workshop on Predictive Intelligence in Medicine (PRIME).
Published version available here.
Authors: Hasib Zunair, Aimon Rahman, Nabeel Mohammed, Joseph Paul Cohen
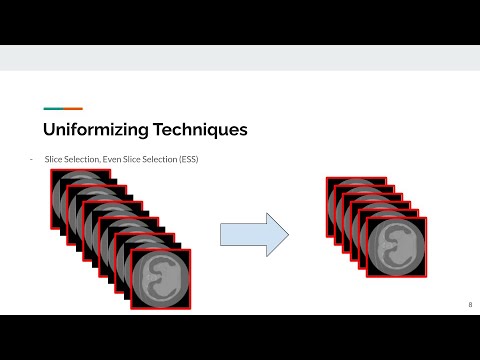
A common approach to medical image analysis on volumet- ric data uses deep 2D convolutional neural networks (CNNs). This is largely attributed to the challenges imposed by the nature of the 3D data: variable volume size, GPU exhaustion during optimization. How- ever, dealing with the individual slices independently in 2D CNNs delib- erately discards the depth information which results in poor performance for the intended task. Therefore, it is important to develop methods that not only overcome the heavy memory and computation requirements but also leverage the 3D information. To this end, we evaluate a set of vol- ume uniformizing methods to address the aforementioned issues. The first method involves sampling information evenly from a subset of the volume. Another method exploits the full geometry of the 3D volume by interpolating over the z-axis. We demonstrate performance improve- ments using controlled ablation studies as well as put this approach to the test on the ImageCLEF Tuberculosis Severity Assessment 2019 bench- mark. We report 73% area under curve (AUC) and binary classification accuracy (ACC) of 67.5% on the test set beating all methods which lever- aged only image information (without using clinical meta-data) achiev- ing 5-th position overall. All codes and models are made available at https://github.com/hasibzunair/uniformizing-3D.
More information about the dataset and task is avaiable at URL.
If you use this code or models in your scientific work, please cite the following paper:
@inproceedings{zunair2020uniformizing,
title={Uniformizing Techniques to Process CT Scans with 3D CNNs for Tuberculosis Prediction},
author={Zunair, Hasib and Rahman, Aimon and Mohammed, Nabeel and Cohen, Joseph Paul},
booktitle={International Workshop on PRedictive Intelligence In MEdicine},
pages={156--168},
year={2020},
organization={Springer}
}Data uniformizing methods
- Ubuntu 14.04
- Python 3.6
- Tensorflow: 2.0.0
- Keras: 2.3.1
You can create the appropriate conda environment by running
conda env create -f environment.yml
- Run notebook in order
others: Contains helper codes to preprocess and visualize samples in dataset.
More details at this link
Zunair, H., Rahman, A., Mohammed, N.: Estimating Severity from CT Scans
of Tuberculosis Patients using 3D Convolutional Nets and Slice Selection. In:
CLEF2019 Working Notes. Volume 2380 of CEUR Workshop Proceedings.,
Lugano, Switzerland, CEUR-WS.org
<http://ceur-ws.org/Vol-2380>(September 9-12 2019) Previous paper published in CEUR-WS. Paper can be found at CLEF Working Notes 2019 under the section ImageCLEF - Multimedia Retrieval in CLEF.
Your driver's license.