-
Notifications
You must be signed in to change notification settings - Fork 3
Home
Geoffrey H. Smith edited this page Jun 28, 2014
·
5 revisions
Thank you for your interest in this project.
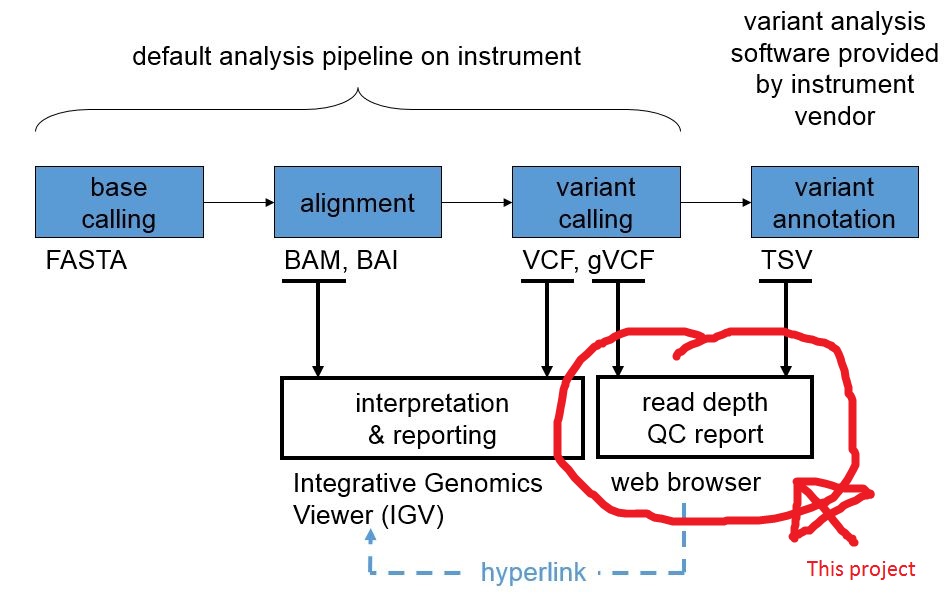
coverageQc generates quality control (QC) reports for next generation sequencing (NGS) data.
There are four inputs:
- A genomic VCF file where the read depths are represented by strings of the form "DP=1234" in the "INFO" field. Note that the genomic VCF file contains entries for both the variant and non-variant positions.
- A BED file which describes the loci that are being reviewed for QC purposes (the "exon" BED file).
- A BED file which describes the coding regions and amplicons associated with the QC loci (the "amplicon" BED file).
- An optional TSV file of annotated variants. The format of this file is vendor-specific.
There is one output:
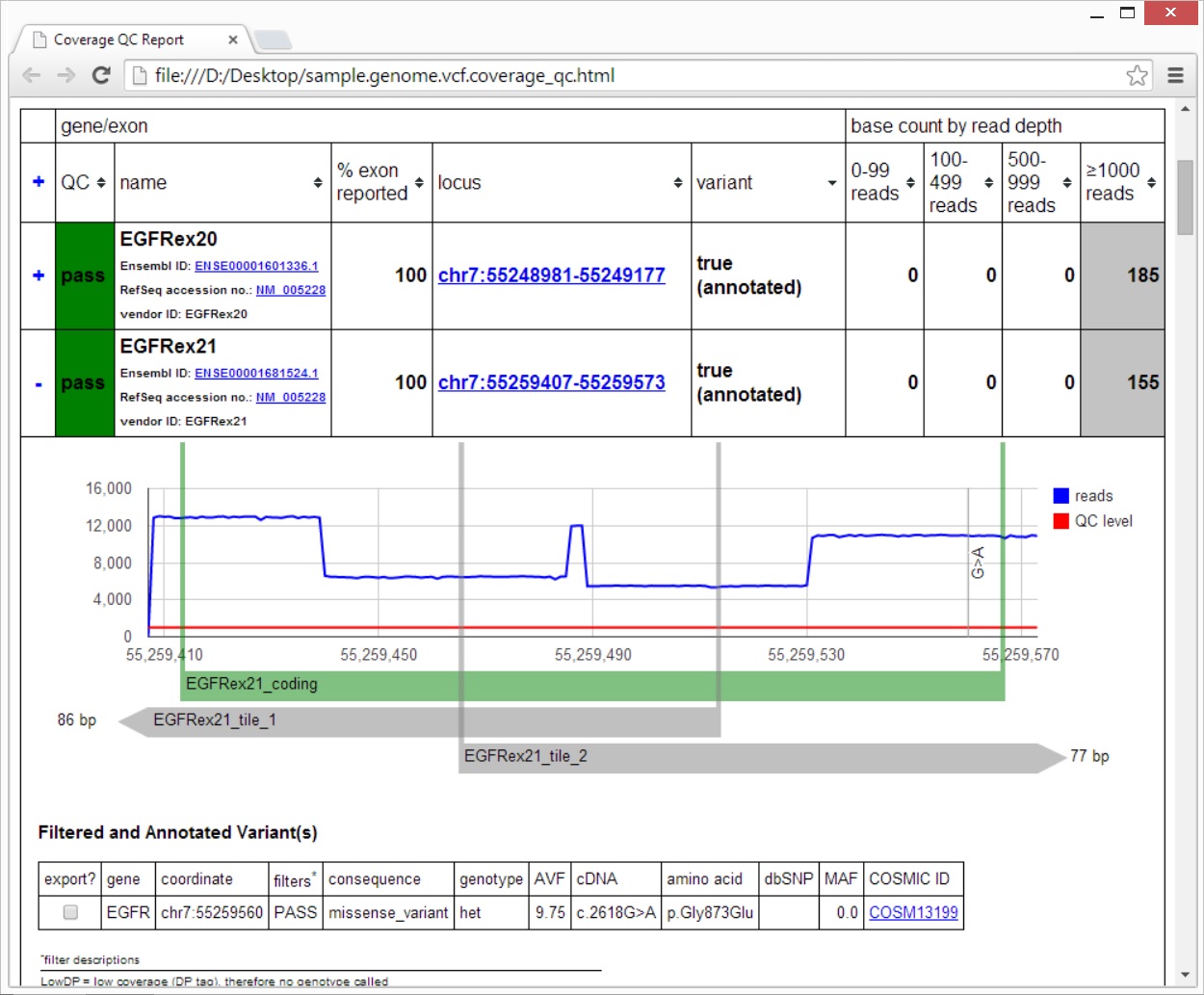
An HTML5 file (see sample report below) that represents the QC report. This file has external dependencies on the jQuery and Google Charts JavaScript APIs. Arbitrary QC rules are applied and each locus in the exon BED file is assigned a QC value of "pass", "warn", or "fail". The read depths are also presented in line-chart format. Links to a locally running IGV session are supported.
This project is implemented as a Java 1.6 application and distributed as a NetBeans 7 project.
Technical point-of-contact: Geoffrey H. Smith, MD (geoffrey.hughes.smith@gmail.com)