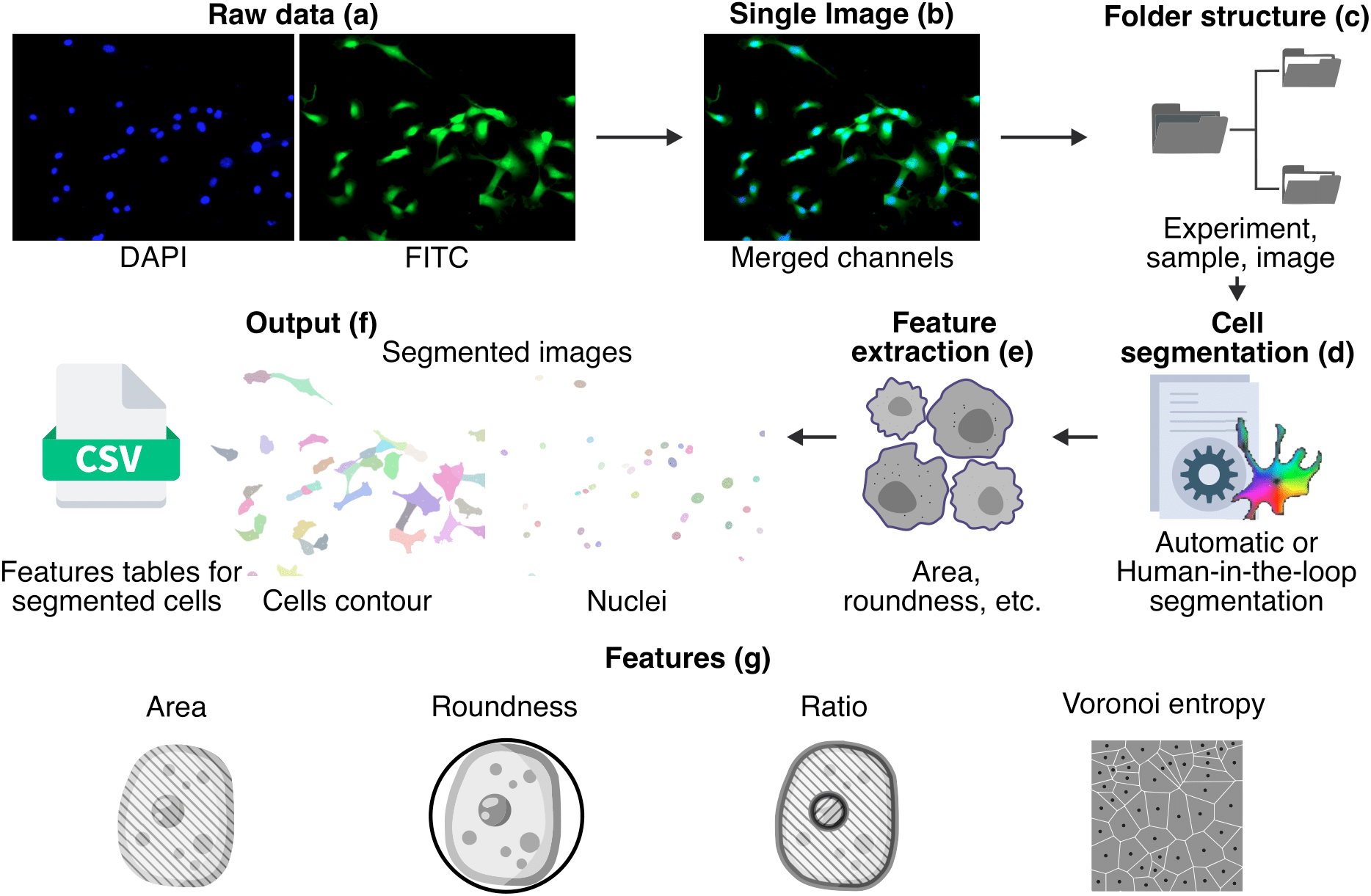
Cellpose plus is a morphological analysis tool that builds on a forked branch of the state-of-the-art image segmentation framework Cellpose.
We add feature extraction algorithms to asses morphological properties of cells and nuclei. This way we achieve a single workflow to study stained cells, from raw images to labeled masks with their corresponding measures.
As the main Cellpose branch continues to grow actively, we aim to keep our forked repository up to date. The latest additions and bug fixes are also present in our repository.
Developed by the InfoChemistry scientific center, part of ITMO University.
We suggest installing our fork using conda and pip (with python>=3.8).
- Install Anaconda.
- Open an
anacondaprompt / command prompt which has conda for python 3 in the path. - For a new environment for CPU only, run:
conda create -n cellpose_plus 'python==3.9' - To activate the new environment, run
conda activate cellpose_plus - For NVIDIA GPUs, run:
pip install torch torchvision --index-url https://download.pytorch.org/whl/cu126
If there are problems with the latest version, we suggest to install CUDA 11.8 - To install the latest PyPi release of Cellpose plus and its dependencies (see setup.py), run:
pip install cellpose-plus[gui]
orpip install cellpose-plusfor a version without GUI. (Optional): To install dependencies, you can use requirements.txt viapip install -r /cellpose_plus/requirements.txt
Linux, Windows and Mac OS are supported for running the code. For running the graphical interface you will need a Mac OS later than Yosemite. At least 8GB of RAM is required to run the software. 16GB-32GB may be required for larger images. The software has been tested on Windows 10, Windows 11, Ubuntu 24.04, Manjaro and limitedly tested on Mac OS.
As a novelty, we contribute with the addition of capabilities to calculate the following metrics:
- Area of subject (𝜇𝑚²).
- Roundness (0.0 - 1.0), having 1.0 for a perfect circle.
- Size ratio between each pair of cell and nucleus.
- Fraction of image covered by cells/nuclei.
- Relative center coordinates.
- Voronoi diagram based on the centers.
- Voronoi entropy, a measure of order/chaos in the cells' positions.
- Convex hull of all objects.
- Continuous symmetry measure (CSM).
In order to obtain metrics from segmented cells, the initial stained images are merged into a single image and organized into sub folders to be processed. A cell segmentation procedure is performed using Cellpose, then we extract the metrics and finally we store the results in the form of images and CSV files.
You can run Cellpose plus in Google Colab with a GPU:
Launching Cellpose plus GUI:
- Launch the command line terminal/Anaconda Prompt:
- Activate respective environments conda activite your_environment (e.g.
conda activate cellpose_plus) - Enter to launch the GUI
python -m cellpose - Now, you can load or drag-drop your desired image for segmentation
Further, we present a usage example:
IMPORTANT: It’s mandatory to set a pixel-to-micrometer(μm) conversion value (μm per pixel), in order to calculate the cells/nuclei area. The input field for this value in the GUI, is named as “Length in μm”. By default this value is automatically acquired if the corresponding metadata file (generated by the microscope after image obtaining) is present in the same folder as the image, and if it shares the same name of the image + ”_Properties.xml”.
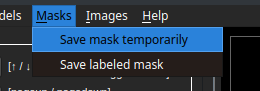
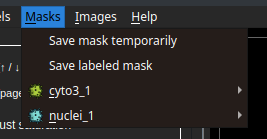
After the segmentation process, possibly including manual editing of the masks, we can save the masks in a folder with the same name as the image and place them in the same location by clicking the "Save labeled mask" button. If we want to calculate metrics for the current segmentation, we can save it as a snapshot by clicking the "Save mask temporarily" button.
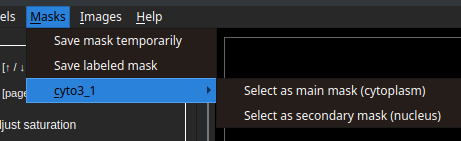
In the image below, we can see a saved snapshot from a mask calculated using a cyto3 model. As it is the first snapshot from this model, the final snapshot name is cyto3_1.
Each snapshot should represent the segmentation of a subject type (cytoplasm or nuclei), to define this, we select one of the options pictured above (main or secondary mask). Here, we see an example of cyto3_1 selected as the main mask and nuclei_1 as the secondary mask.
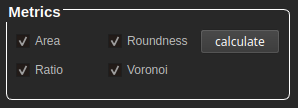
At the bottom of the GUI, we find the metrics panel with the following options:
Area and roundness are clickable when having a snapshot selected as primary. If there is a primary and a secondary snapshot available, the values are calculated separately per subject (cells and/or nuclei).
Ratio and Voronoi are clickable when having a primary and a secondary snapshot selected. To obtain results, both snapshots are necessary.
After clicking "calculate" it will take a few moments until we get a folder with the same name as the source image, containing the result values in .csv and .png formats. For extra feedback about the processes and alerts, we suggest to stay pending of the python shell.
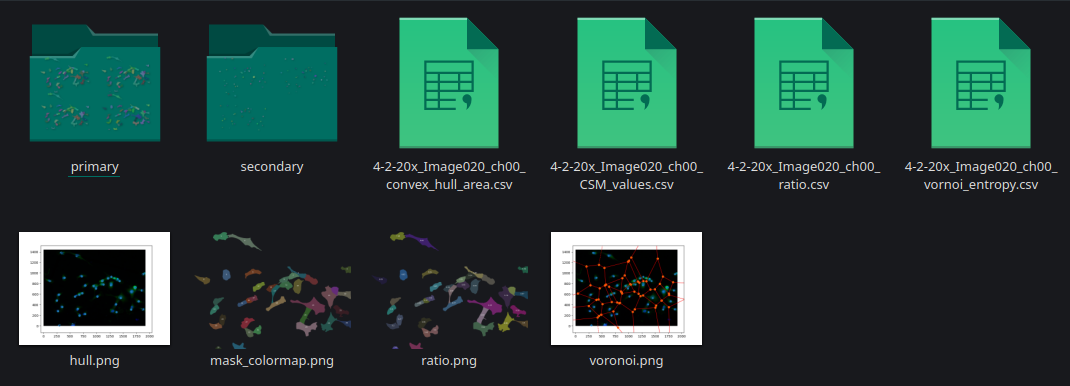
The resulting directory consists of "primary" and "secondary" folders with individual results per snapshot. For example: when analyzing an image of cells, after segmentation the directory will contain the following folders
-
Primary: Contains the area and roundness of cells (where, cell = nuclei and cytoplasm) as .png masks and two .csv files.
-
file_name_Center.csv = A csv file containing the center coordinates of the cell in [X,Y] format.
-
file_name_size_roundness.csv = A csv file containing the area of the cell and its roundness in [area,roundness] format.
-
-
Secondary -> contains the area and roundness of the nuclei as .png masks and a .csv file.
- file_name_size_roundness.csv = A csv file containing the area of the nuclei and roundness in [area,roundness] format.
In instances where ratio and Voronoi entropy are selected. The results of the metrics are saved in the parent directory as the image.
-
Continous symmetry measure (CSM): The symmetry of cell is stored in file_name_CSM_values.csv format as [cell id, CSM_metric_value].
-
Ratio: The ratio of cell to nuclei is stored in file_name_ratio.csv format as [cell id, nuclei id, ratio].
-
Voronoi entropy: The vornoir entropy of the entire image is stored as a single value in file_name_vornoi_entropy.csv and its respective image as voronoi.png
For features provided by the basic Cellpose, such as image restoration, segmentation settings and mask editing, we encourage you to refer to the original Cellpose documentation.
If you find our project helpful, use the following bibtex to reference our paper.
@article{huaman2025cellpose+,
author = {Huaman, Israel A. and Ghorabe, Fares D. E. and Chumakova, Sofya S. and Pisarenko, Alexandra A. and Dudaev, Alexey E. and Volova, Tatiana G. and Ryltseva, Galina A. and Ulasevich, Sviatlana A. and Shishatskaya, Ekaterina I. and Skorb, Ekaterina V. and Zun, Pavel S.},
title = {Cellpose+, a Morphological Analysis Tool for Feature Extraction of Stained Cell Images},
journal = {Advanced Intelligent Discovery},
volume = {n/a},
number = {n/a},
pages = {202500005},
keywords = {bioimaging, image analysis, image segmentation, microscopy},
doi = {https://doi.org/10.1002/aidi.202500005},
url = {https://advanced.onlinelibrary.wiley.com/doi/abs/10.1002/aidi.202500005},
eprint = {https://advanced.onlinelibrary.wiley.com/doi/pdf/10.1002/aidi.202500005},
abstract = {Advanced image segmentation and processing tools present an opportunity to study cell processes and their dynamics. However, image analysis is often routine and time-consuming. Nowadays, alternative data-driven approaches using deep learning are potentially offering automatized, accurate, and fast image analysis. In this paper, we extend the applications of Cellpose, a state-of-the-art cell segmentation framework, with feature extraction capabilities to assess morphological characteristics. We also introduce a dataset of 4′,6-diamidino-2-phenylindole and fluorescein isothiocyanate stained cells to which our new method is applied.}
}
As we work over Cellpose, we ask you to also cite the Cellpose paper.