-
Notifications
You must be signed in to change notification settings - Fork 3
UI_Video
Tag video frames and trials with metadata and synchronize videos with electrophysiologic data.
Make sure to check the setup tutorial
nigeLab.libs.VidScorer(blockObj.Cameras)
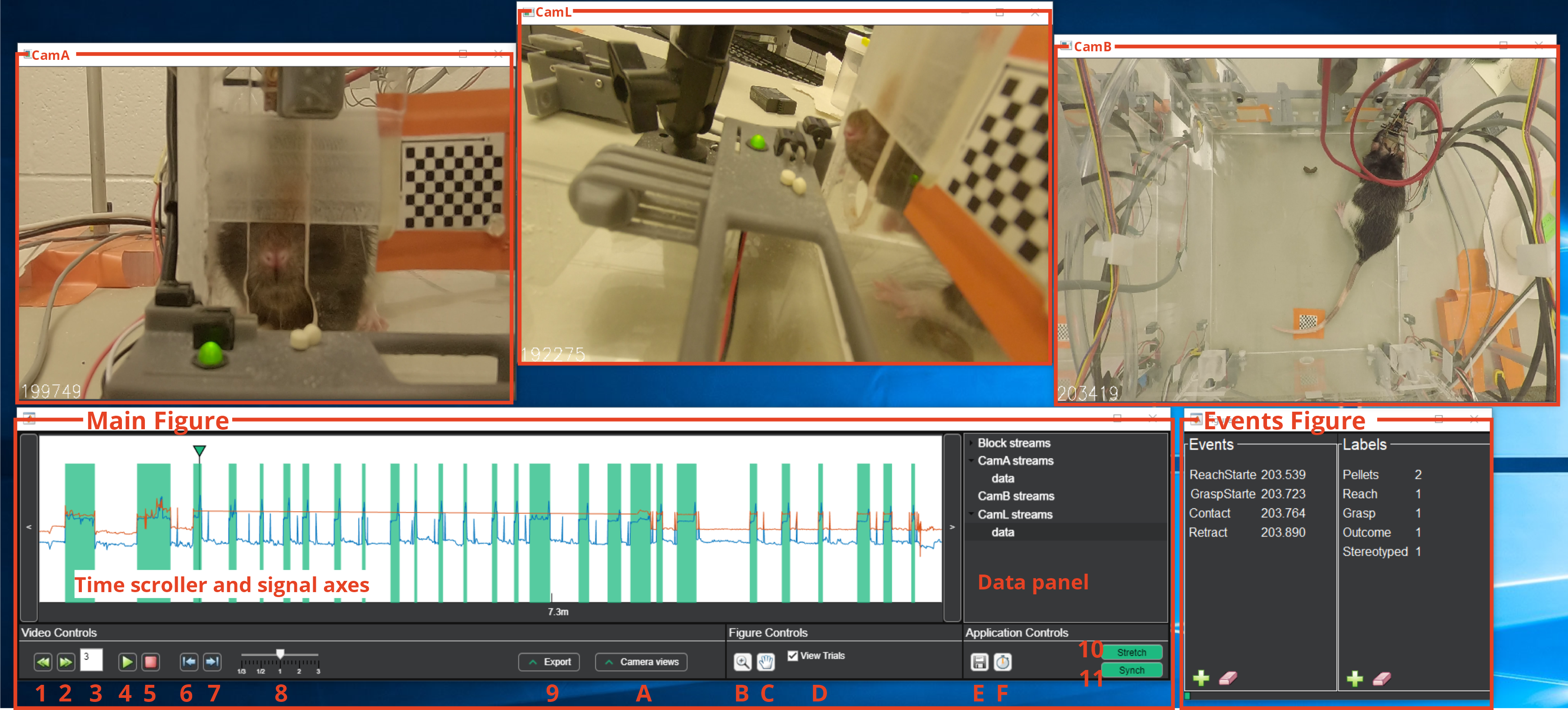
Wait until loading is complete. Three windows should open up:
- The Main Figure with the controls, the plots and the data tree.
- A window displaying a video frame. This will have the same name as the nigelCamera it is sourced from (here CamA).
- The Events Figure with an Event and a Labels panel.
Most of the action takes place on the Main Figure: here you can scroll through the video by clicking anywhere on the signal axes where trials (green rectangles, if not masked, yellow if masked) and streams (here blue and orange traces) are visualized. A time indicator shows the time of the current displayed frames. On the Data Panel are shown all the data sources available to plot: Block streams contains all the auxiliary data recorded together with the electrophysiological signal while video streams (here CamA, CamB and CamL) contains all the auxiliary data extracted from the videos or externally provided. While Block Streams are linked to the BlockObj and thus are not editable, Video Streams can be added and removed from the contextual menu (See below).
A description of the available controls and their function is provided below:
- Previous trial
- Next trial
- Shows the current trial number. If modified jumps to the provided trial.
- Play/Stop Buttons | Control the video
- Start/Stops buffering
- Previous frame
- Next frame
- Adjust playback speed
- Opens the export panel
- A. Opens the Camera Views panel
- B. Toggles zoom along x-axis on the signal axis. From the right-click menu is possible to zoom out to the default zoom.
- C. Toggles pan along x-axis along x-axis.
- D. Show/hide trials on the signal axis
- E. Toggles a 5-minutes autosave
- F. Starts performance monitor. This function makes a very basic estimate of how much time will be needed to complete scoring for the current video and at the current speed. When deactivated it displays the results in the command window.
- Stretch mode. Together with the Synch mode it is used to align the videos with the ePhys data.
- Synch mode. Together with the Stretch mode it is used to align the videos with the ePhys data.
The most used controls can also be accessed by keyboard hotkeys, defined in the file +workflow\defaultVideoScoringHotkey.m.
Pressing h prints a formatted description of the current hotkeys configuration.
Hotkeys can also be used to add events to the current frame or labels the the current trial.
Streams can be extracted from the video or can be imported externally and are used to align videos and electrophysiologic data. The video extracted streams consist of normalized median luminance values computed and normalized over two user defined ROIs. Externally imported streams have to be saved in a matlab readable format and either provide a time vector or be consistent with the video length. To add a stream from videos:
- In the data panel, right-click on a camera you want to add a stream to and select “add stream from video”
- Two new video frames will open in succession— The first one to create a ROI box around a signal source (e.g. an LED whose driving signal was coregisterd with the ePhys) and the second one to create a larger box as a normalization factor
- To draw the ROI box click and drag. Press
spaceto confirm. - A sketchy progress bar will appear on screen. Wait until completion or stop the execution by pressing
Esc - The new video stream will appear in the data panel. It can be deleted or visualized using its context menu.
To add an external stream:
- In the data panel, right-click on a camera you want to add a stream to and select “add external stream”
- A file selector will pop up where you can select a matlab-format file containing the desired stream
- If it has two variables in it the system will assume one is data and the other is the time vector, which has to be called
timeort - If only one variable is present, the user will be prompted to either select another mat-file containing the time vector or to use the same time basis as the video.
- If it has two variables in it the system will assume one is data and the other is the time vector, which has to be called
Through the stretch and synch functions videos and ePhys data can be aligned.
- Plot the video-streams to be used for alignment using their context-menu
- Select Synch by clicking the corresponding button.
- Drag the video streams to overlap them with the rising/falling edges of the Trials overlay (or some other feature e.g. a Block stream)
- Zoom in to increase the accuracy of the match particularly at the beginning of the recording
- If the synchronization is lost over the course of the recording despite aligning at the beginning, there is a mismatch in the camera's and ePhys' clocks.
- Zoom in at the end of the recording and select Stretch to alter camera timing to fit the digital stream.
- Drag the desired signal until it is aligned to the selected feature.
- Return to the beginning of the recording and select **Sync **to realign after the stretch
- Rinse and repeat until satisfaction (two time is usually enough)
- Clicking the plus sign under "Events" in the Events figure will tag a frame of interest
- Clicking the plus sign under "Labels" in the Events figure will tag the individual trial
- Clicking the eraser sign will delete the currently selected Event or Label
- Events and labels are shown only while displaying the trial they are on
- Events and Labels can be modified or deleted from the context menu
- After progress is saved, the tags and their timestamps can be accessed in
blockObj.Events
A method is provided to automatically parse events from rising and falling edges of a digital input stream
- As a default, the configuration file
+defaults\Event.mis set so as to catch the start and stop of a trial based on a digital input a+d respectively call these two eventsBeginTrialandEndTrial. - For each event a structure is defined, whose name corresponds to the given name of the event. Inside this structure some parameters are defined:
- Source -> [string or char array], the name of the Block's stream used to parse the event
blockObj.Streams.DigIO - DetectionType -> [
'Rising'or'Falling'], it defines the type of edge associated with this event. - Debounce -> [double],
- Tag -> [string or char array], a short name.
- Source -> [string or char array], the name of the Block's stream used to parse the event
Example:
pars.BeginTrial = struct('Source','trial-running',...
`'DetectionType','Rising',...
`'Debounce',0,...
`'Tag','BTrial');blockObj.doEventDetection
Trials can be masked if needed. A mask can be set with blockObj.setTrialMask(MaskingArray) method. MaskingArray can either be a logical vector the same length as number of trials, a numeric vector with trials' indexes or it can be omitted (or an empty vector) to restore default.
When masked a trial appears as a yellow rectangle in the VideoScorer and cannot be jumped to with shortcuts or buttons.
A method is provided to parse the video names and automatically match them to the correct block. The are several ways to organize videos compatibly with this feature:
In this case all the metadata will be sourced from the folders name, thus not requiring strict naming of each individual video file. The video files still need to be named in a way that makes them sortable, but the actual naming can be arbitrary (e.g. the GoPro naming scheme). An ordering function should be provided to match this naming scheme. Scenario 1: One folder per animal, one folder per camera
Videos
|__ Animal1
| |__ Animal1_Therapy1_Date_CamA
| | |__ Video1.mp4
| | |__ ...
| | |__ VideoZ.mp4
| |
| |__ Animal1_TherapyX_Date_CamY
| |__ Video1.mp4
| |__ VideoZ.mp4
|
|__ AnimalN
|__ AnimalN_Therapy1_Date_CamA
Scenario 2: All animals, one folder per camera
Videos
|__ Animal1_Therapy1_Date_CamY
| |__ Video1.mp4
| |__ ...
| |__ VideoZ.mp4
|
|__ AnimalN_TherapyX_Date_CamY
|__ Video1.mp4
|__ ...
|__ VideoZ.mp4
In this case all the metadata will be sourced from the video names, which will require strict naming with also an ordering field.
Scenario 3: All together
Videos
|__ Animal1_Therapy1_Date_CamA_Video1.mp4
|__ Animal1_Therapy1_Date_CamA_Video2.mp4
|__ Animal1_Therapy2_Date_CamA_Video1.mp4
|__ Animal1_Therapy2_Date_CamA_Video2.mp4
|__ ...
|__ AnimalN_TherapyX_Date_CamY_VideoZ.mp4
- Set
pars.HasVideo = true; - Input the top folder containing the videos under
pars.VidFilePath(in the examples above it would beVideos) - In
pars.UniqueKey.varsinsert the name of Block's metadata variables that will be concatenated and used to match the video names -
pars.NamingConventionworks to save the metadata from the video name in a similar way to what is done with parsing of the initial tank file name set-up - If a custom sorting function is needed (Scenario 1 and 2) input it as a handle in
pars.CustomSort - If the videos are sorted depending on a piece of metadata derived from the naming (Scenario 3), set the corresponding field in
pars.GroupingVar
Example, scenario 1 and 2
pars.UniqueKey.vars = {'AnimalID','Therapy','Date'};
pars.UniqueKey.cat = '_';
pars.NamingConvention={'$AnimalID','$Therapy','$Date','$CameraID'};
pars.FileExt = '.MP4';
pars.Delimiter = '_';
pars.SpecialMeta = struct;
pars.SpecialMeta.SpecialVars = {'VideoID'};
pars.SpecialMeta.VideoID.cat = '-';
pars.SpecialMeta.VideoID.vars = {'AnimalID','Phase','RecDate','CameraID'};
pars.GroupingVar = 'CameraID';
pars.IncrementingVar = '';
pars.CustomSort = @nigeLab.utils.orderGoProVideos;Example, scenario 3. Only reporting the differences from above
pars.NamingConvention={'$AnimalID','$Phase','$RecDate','$CameraID','$Counter'};
pars.IncrementingVar = 'Counter';
pars.CustomSort = [];
- Check
blockObj.Camerasfor correct assignment. There should be one nigelCamera object for each different camera associated with the block. Each object should have a struct-array under the Meta property with the number of elements matching the number of videos recorded by this camera.
path = fullfile('K:\Rat\Video\UnilateralReach\Experimental_Folder\Animal_Folder','R20-99_2020_1_11_0_Left_A_0.mp4');
camObj = nigeLab.libs.nigelCamera(blockObj,{path});or with multiple video files for the block
path = {'K:\Rat\Video\UnilateralReach\Experimental_Folder\Animal_Folder\R20-99_2020_1_11_0_Left_A_0.mp4',
K:\Rat\Video\UnilateralReach\Experimental_Folder\Animal_Folder\R20-99_2020_1_11_0_Left_A_1.mp4'};
camObj = nigeLab.libs.nigelCamera(blockObj, path);