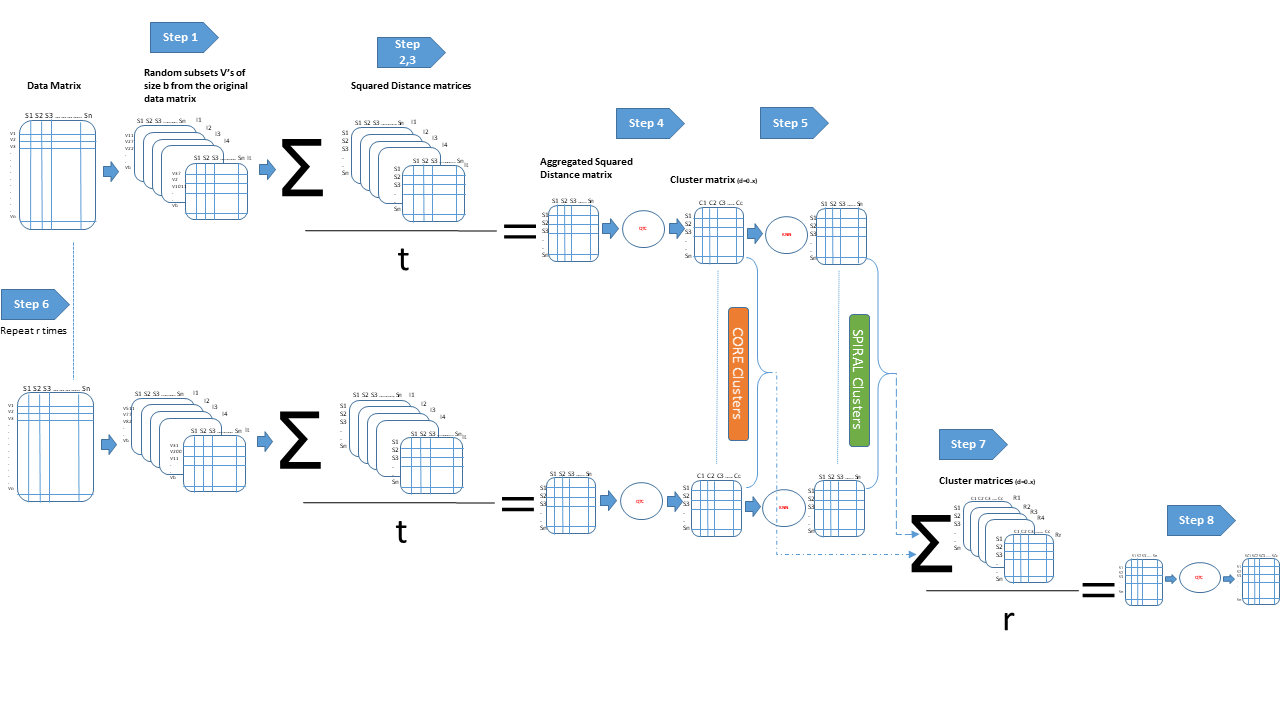
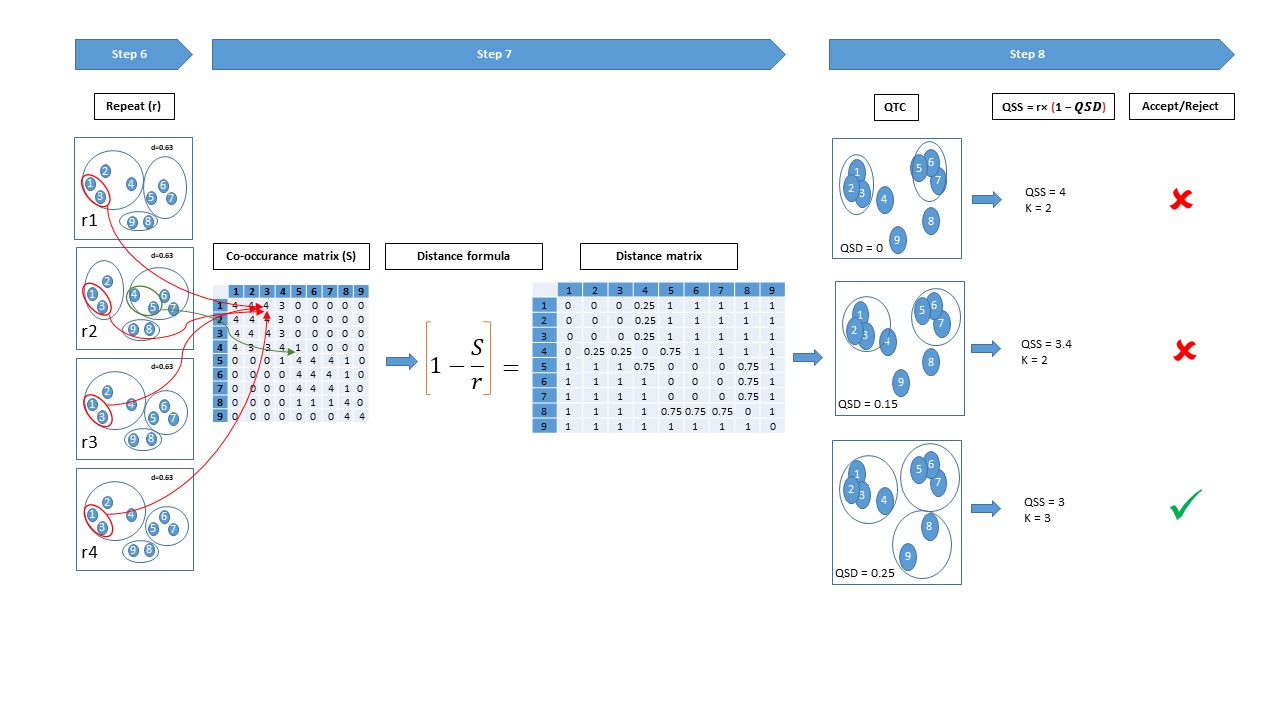
This is an implementation of Systematic Random forest Integration to Qualitative threshold (SRIQ) clustering. SRIQ clusters by finding a core clusters of highly correlated observations, then spiralling out from them to create bigger clusters. SRIQ evaluates clustering solution stability on its own and won't need user input for what number of cluster solutions to be evaluated.
https://www.sciencedirect.com/science/article/pii/S2001037022001118
Step 1: Data Pre-process
e.g., Input File (FPKM) format
Gene TCGA-05-4384-01A TCGA-05-4390-01A TCGA-05-4396-01A TCGA-05-4405-01A TCGA-05-4410-01A TCGA-05-4415-01A TCGA-05-4417-01A TCGA-05-4424-01A TCGA-05-4425-01A TCGA-05-4427-01A TCGA-05-4433-01A
ENSG00000242268.2 0.12364017 0 0.148772639 0 0 0 0 0 0 0 0.044008461
ENSG00000270112.3 0 0.005866919 0.006880892 0.006391225 0 0 0 0 0 0.004996734 0
ENSG00000167578.15 4.04324606 2.043595153 2.021171586 3.11505239 5.089686463 1.266061817 3.569842733 2.468971417 2.294267819 1.805431595 3.714282814
ENSG00000273842.1 0 0 0 0 0 0 0 0 0 0 0
ENSG00000078237.5 4.917249583 4.826403272 3.284514027 4.132612814 4.658567151 8.91755157 8.215148814 4.059616218 6.055433683 3.670736747 4.175781332
ENSG00000146083.10 21.96680966 10.92637933 15.83184449 10.0769604 9.476091577 9.067037219 9.865930315 9.246771338 8.79136018 11.96198371 15.23656895
ENSG00000225275.4 0 0 0 0 0 0 0 0 0 0 0
ENSG00000158486.12 0.522247482 0.134452442 0.440119056 0.430659602 1.206752222 0.069414068 0.276998364 0.70092943 0.041085547 0.205093223 0.41146146
ENSG00000198242.12 120.6622689 176.9369348 113.4584536 103.3924893 117.0184252 248.6506293 161.1558253 217.6540163 245.8388782 144.1747257 109.1065434To preprocess the data, navigate to the folder in which the SRIQPreprocess.jar file exist and run following command:
java -jar SRIQPreprocess.jar path-to/datafile.txtoutput: a file with name ending with "_mc_log2_nz" will be in the same path folder as input file folder.
Gene TCGA-05-4384-01A TCGA-05-4390-01A TCGA-05-4396-01A TCGA-05-4405-01A TCGA-05-4410-01A TCGA-05-4415-01A TCGA-05-4417-01A TCGA-05-4424-01A TCGA-05-4425-01A TCGA-05-4427-01A TCGA-05-4433-01A
ENSG00000242268.2 0 0 0 0 0 0 0 0 0 0 0
ENSG00000167578.15 0.08146491 -0.90293956 -0.9188573 -0.29479265 0.41352773 -1.5937011 -0.098188564 -0.630139 -0.7360152 -1.0817053 -0.040965408
ENSG00000078237.5 0.22821039 0.20130733 -0.35396126 -0.022586951 0.15024513 1.0870064 0.9686455 -0.04829784 0.52858907 -0.19357152 -0.007595019
ENSG00000146083.10 0.76856166 -0.23894827 0.29606566 -0.35570318 -0.44439968 -0.5080606 -0.38623673 -0.4797421 -0.55260533 -0.10830704 0.24077445
ENSG00000158486.12 0 0 0 0 0.27112943 0 0 0 0 0 0
ENSG00000198242.12 -0.24240795 0.30985266 -0.33121842 -0.46525115 -0.28664684 0.8007375 0.17507376 0.608654 0.7843305 0.014435649 -0.38764498
ENSG00000259883.1 0 0 0 0 0 0 0 0 0 0 0
ENSG00000231981.3 0 0 0 0 0 0 0 0 0 0 0
ENSG00000134108.11 0.6123477 0.07525928 0.5484164 0.2805461 -0.23346047 -0.43867207 0.23890966 0.29994708 0.33914524 0.7241365 0.24598958
ENSG00000172137.17 0 0 0 0 0 0 0 0 0 0 0.38148433
ENSG00000167700.7 0.34559175 2.1602454 0.6777301 -0.5876767 -0.9264774 -0.73489106 0.085830145 -1.2372832 -0.8909387 -1.6027871 -0.36108896
ENSG00000234943.2 0 0 0 0 0 0 0 0 0 0 0
ENSG00000060642.9 0.863422 -0.8631396 -0.3552939 -0.20815371 0.1698215 -0.13628596 -0.43969843 -0.7006184 -0.24632233 -0.4998986 -0.24191558
ENSG00000231105.1 0 0 0 0 0 0 0 0 0 0 0
ENSG00000182141.8 0.19416559 -0.21492973 -0.54135746 0.7849283 0.52448696 -0.54135746 -0.45162266 1.2665613 0.027005259 0.6991696 -0.54135746Step 2: Edit the test.properties file, inFileName should be the output file from SRIQPreprocess (step 1)
#testing
studyName=LU_LUAD_SRIQ
studyPath=/home/Researcher/SRIQ/software/data/
inFileName=GeneData_AC_fpkm_mc_log2_nz
outPath=/home/Researcher/SRIQ/software/output/
distCutOff=0.8, 0.79, 0.78, 0.77, 0.76, 0.75, 0.74, 0.73, 0.72, 0.71, 0.7, 0.69, 0.68, 0.67, 0.66, 0.65, 0.64, 0.63, 0.62, 0.61, 0.6, 0.59, 0.58, 0.57, 0.56, 0.55, 0.54, 0.53, 0.52
permutations=10000
iterations=10
spiral=TRUE
minClusterSize=0
minBagSize=1200
method=PEARSONStep 3: SRIQ clustering
To run SRIQ, navigate to the folder in which the SRIQS.jar file exist and run following command:
java -jar -Xmx8g SRIQS.jar VRLA path-to/test.propertiesoutput path: e.g.,..\LUAD_SRIQ\LUAD_2021_FPKM_test_10000itr_1200var_10r\10000\QC_Spiral(false)\
Step 4: Select cluster solution from the image e.g., ..._Clusters_Frequencies.pdf
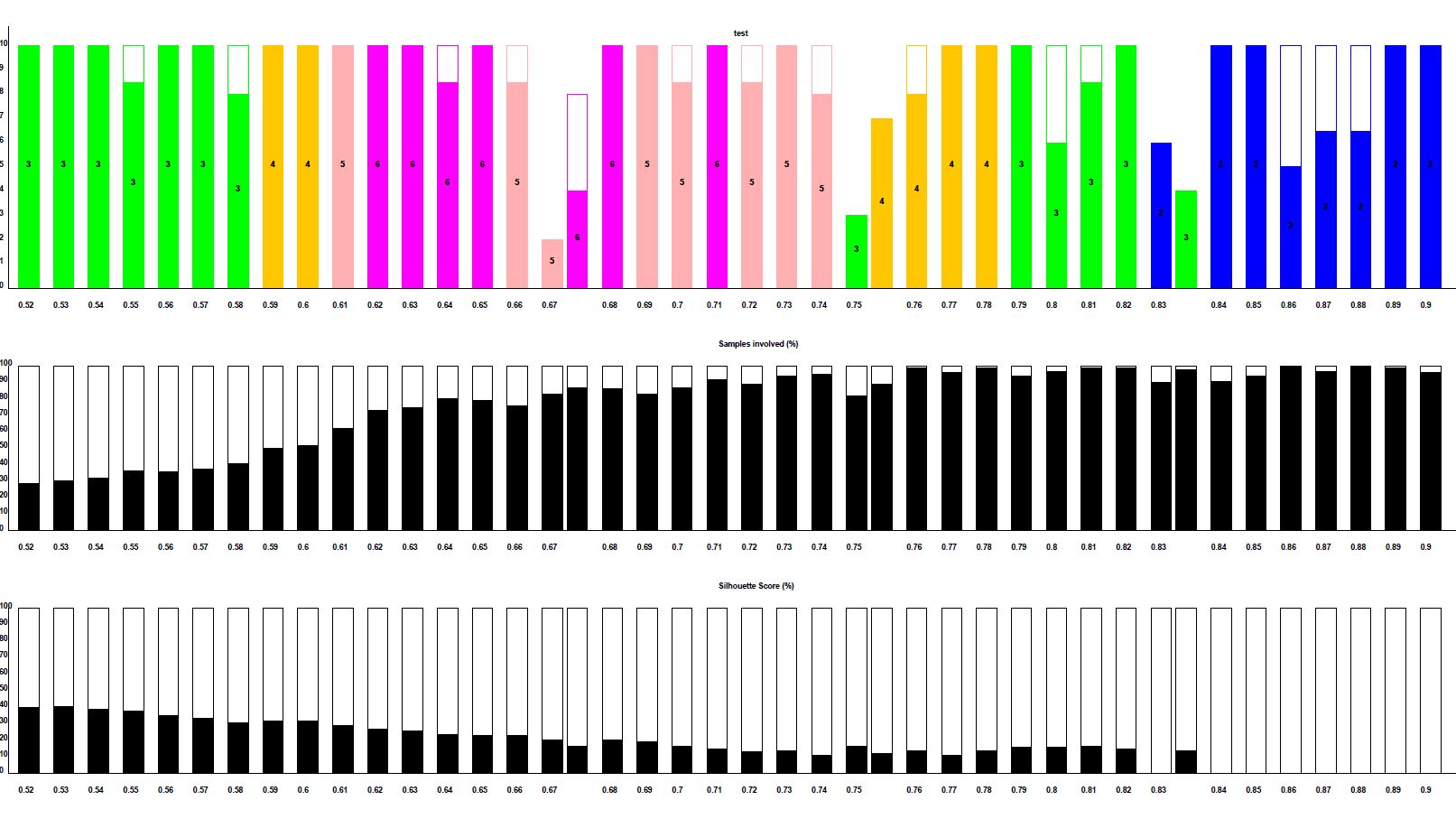
Step 5: Extract Clusters data
To extract clusters data, navigate to the folder in which the SRIQS.jar file exist and run following command:
java -jar <path-to/SRIQS.jar> EXTRACT <path-to/test.properties> <spiral (true or false)> <diameter> <no. of clusters> <log2_transformed_gex_file>
e.g., java -jar SRIQS.jar EXTRACT "F:/test/LUAD_test/test.properties" false 0.63 6 "F:/test/LUAD_test/newFiltered_35k.txt"output path: e.g., ...\LUAD_SRIQ\LUAD_2021_FPKM_test_10000itr_1200var_10r\10000\QC_Spiral(false)\Results_log_0.63_6\
Step 6: SAMDEG (Differentially Expressed Genes Analysis)
To run SAMDEG, navigate to the folder in which the SRIQS.jar file exist and run following command:
java -jar <path-to/SRIQS.jar> SAMDEG <path-to/test.properties> <spiral (true or false)> <diameter> <no. of clusters> <q-value> <fold-change> <log2_transformed_gex_file>
e.g., java -jar -Xmx8g SRIQS.jar SAMDEG "F:/test/LUAD_test/test.properties" false 0.63 6 0 2 "F:/test/LUAD_test/newFiltered_35k.txt"output path: e.g., ...\LUAD_SRIQ\LUAD_2021_FPKM_test_10000itr_1200var_10r\10000\QC_Spiral(false)\Results_log_0.63_6\
To install this repository simply create a folder and clone the repository:
git clone https://github.com/StaafLab/SRIQ
cd SRIQ
pip install -r requirements.txtTo run the pipeline, start the jupyter notebook file and follow the instructions within the pipeline. It's recommended to use virtual environments when running the pipeline in order to avoid conflicting packages.
jupyter notebook analysis_pipelineTo run SRIQ, navigate to the folder in which the SRIQ.jar file exist and run following command:
java -jar SRIQ.jar path-to/test.propertiesAlternative to run SAMDEG, navigate to the folder in which the SAMDEG.jar file exist and run following command:
java -jar <path-to/SAMDEG.jar> <path-to/test.properties> <spiral (true or false)> <diameter> <no. of clusters> <q-value> <fold-change> <log2_transformed_gex_file>
e.g., java -jar SAMDEG.jar "F:/test/LUAD_test/test.properties" false 0.63 6 0 2 "F:/test/LUAD_test/newFiltered_35k.txt"The project provide following features:
- SRIQ-clustering
- Pre-clustering data normalization
- Silhoutte-plot analysis
- UMAP
- Differential gene expression with SAM
- Gene enrichment analysis with EnrichR against customizable databases
- Visualization of single or multiple genes across clusters
Your data, to be clustered, should be a tab separated .txt file and look like this:
For SRIQ to accept the data to be clustered, the file has to be in following format:
| Gene | Sample1 | Sample2 | ... | SampleN |
|---|---|---|---|---|
| Genename 1 | val1 | val2 | ... | valN |
| ... | ... | ... | ... | ... |
| Genename M | val1 | val2 | ... | valN |
For clustering, expression data should be:
- Off-set by 0.1 OR set all values < 1 set to 1
- Log2transformed
- Median-centered
For SAM-analysis:
- Off-set by 0.1 OR all values < 1 set to 1
- Log2transformed
Before running SRIQ, test.properties file need to be correctly configured. The following lines needs to be correct otherwise SRIQ won't start.
studyName: Desired output name for project
studyPath: Folderpath to expression data
inFileName: expression file. exclude '.txt' from file name
outPath: Folderpath for SRIQ output
The enrichR module assumes that gene names are in the form of gene symbols. I have implemented mygene api, set the variable 'scopes' as the format of your gene names.
For python packages run following command:
pip install -r requirements.txtTo run SRIQ and SAM analysis, java is needed on your system.
Copyright (C) 2021 Srinivas Veerla
This program is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License as published by the Free Software Foundation, either version 3 of the License, or (at your option) any later version.
This program is distributed in the hope that it will be useful, but WITHOUT ANY WARRANTY; without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. See the GNU General Public License for more details.
You should have received a copy of the GNU General Public License along with this program. If not, see https://www.gnu.org/licenses/.