-
Notifications
You must be signed in to change notification settings - Fork 1
Add CMS_TTB_8TEV_2D grids #8
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
base: master
Are you sure you want to change the base?
Conversation
|
Result for |
|
Result for the |
…pl.lz4,CMS_TTB_8TEV_2D_TTM_TTRAP.pineappl.lz4: convert to Git LFS
 enocera
left a comment
enocera
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
May I suggest to change the y-axis label in the first inset of all of the plots in order to clarify that the distribution is double differential? Example (for the first plot):
dsigma/dyt -> dsigma/dytdpTt
|
@enocera The previous commit makes all distributions two dimensional. Results: |
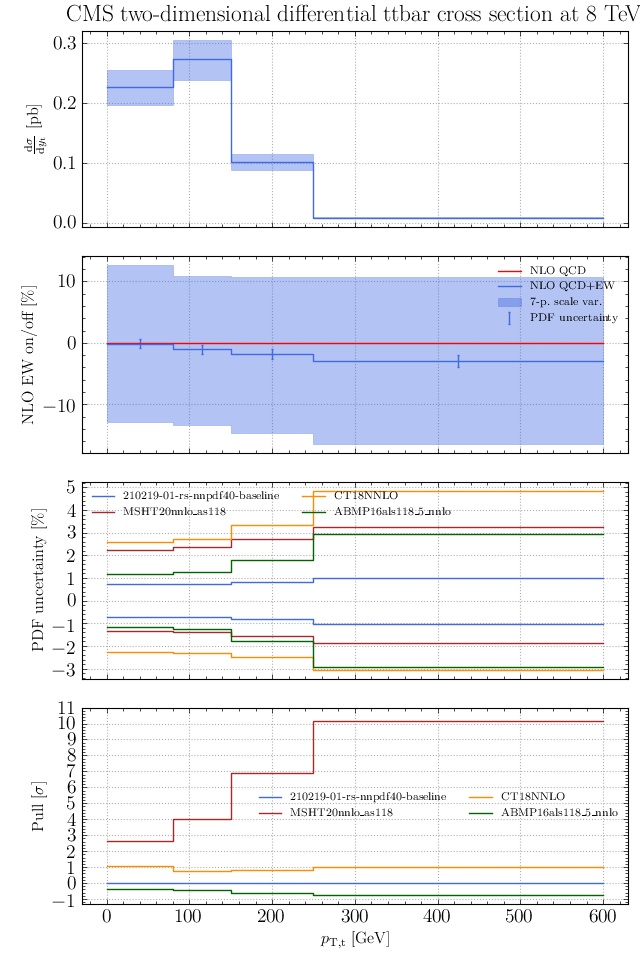
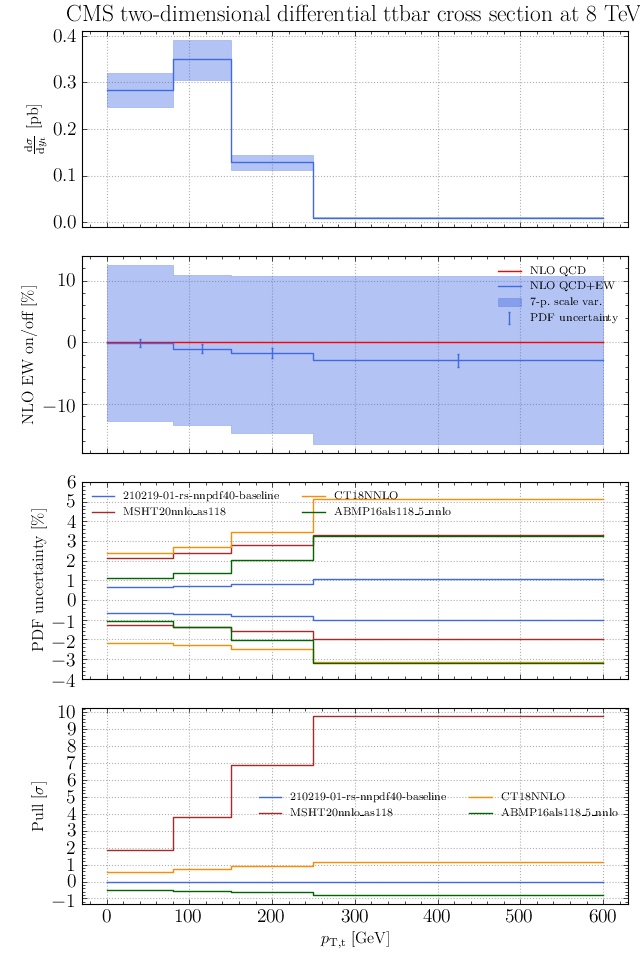
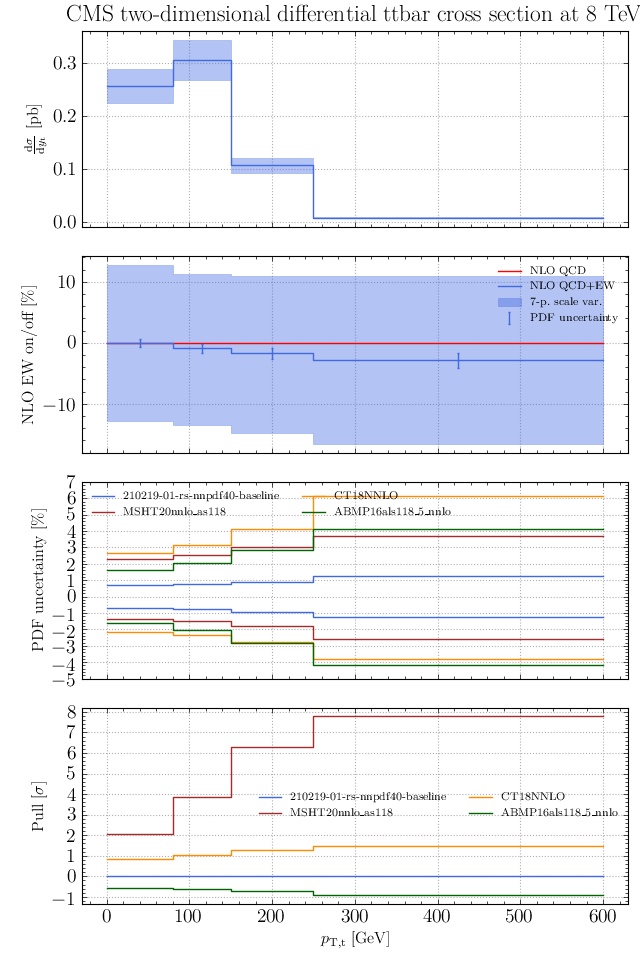
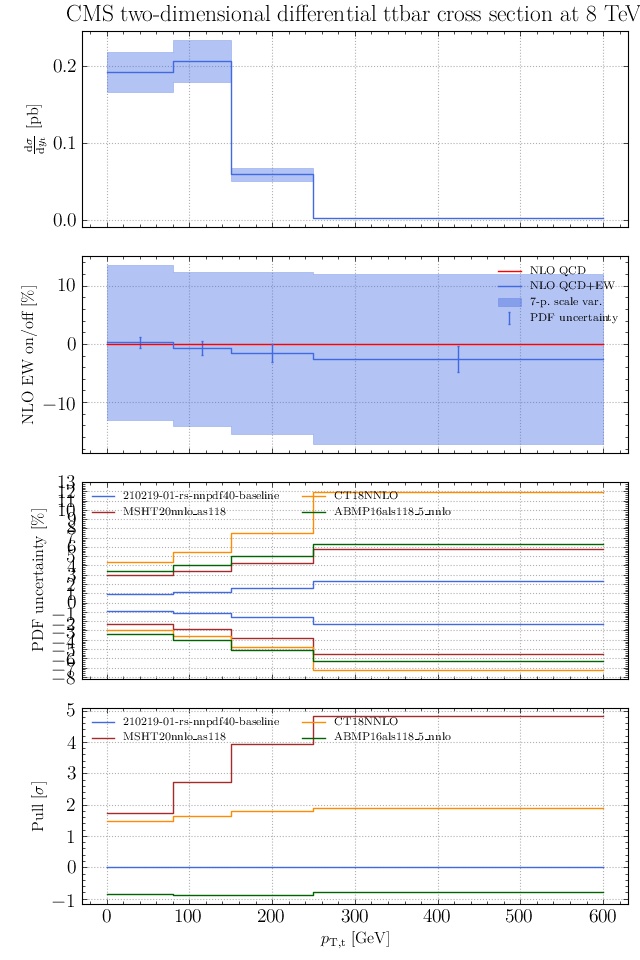
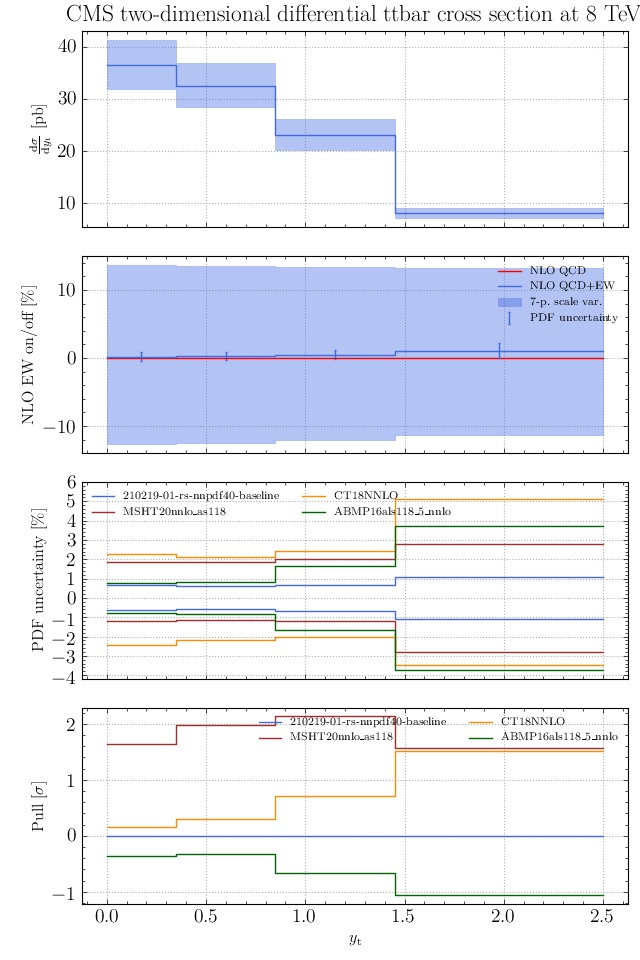
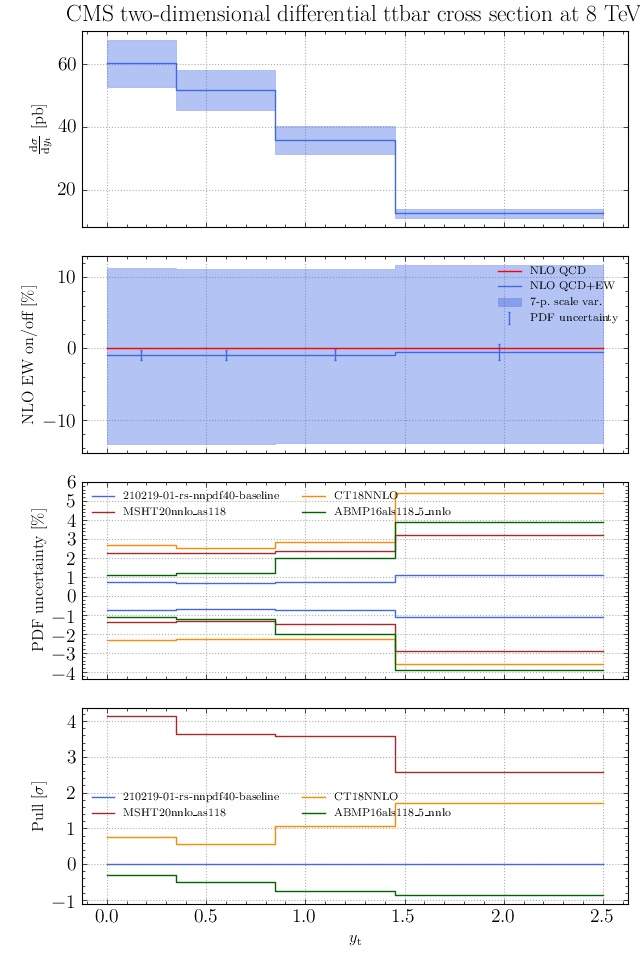
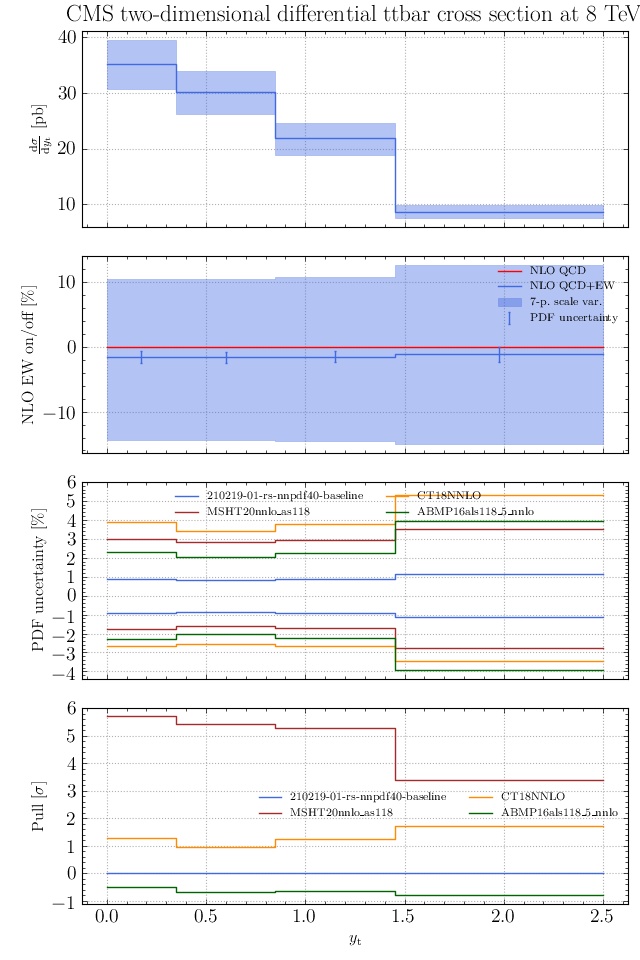
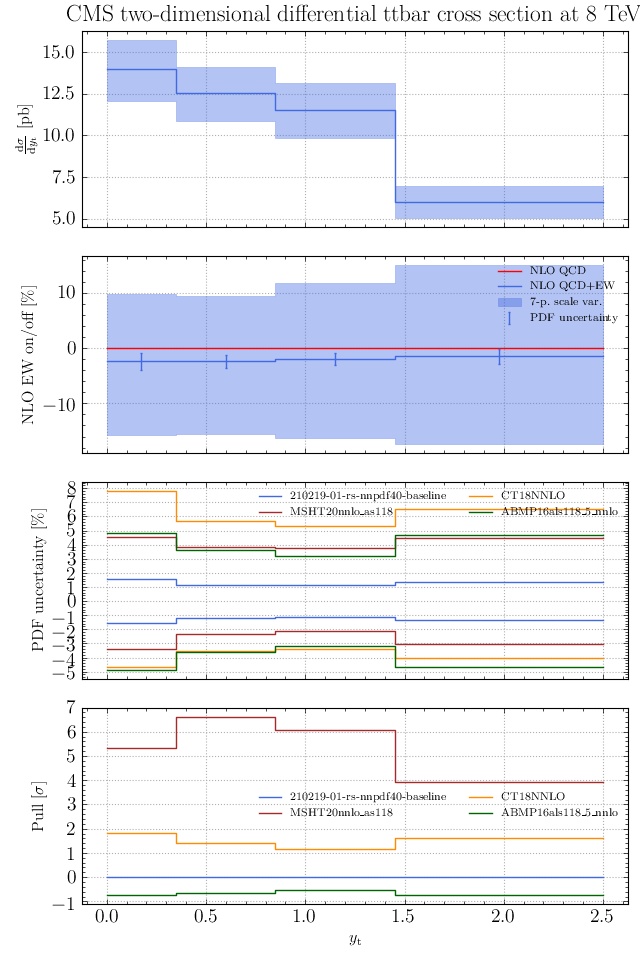
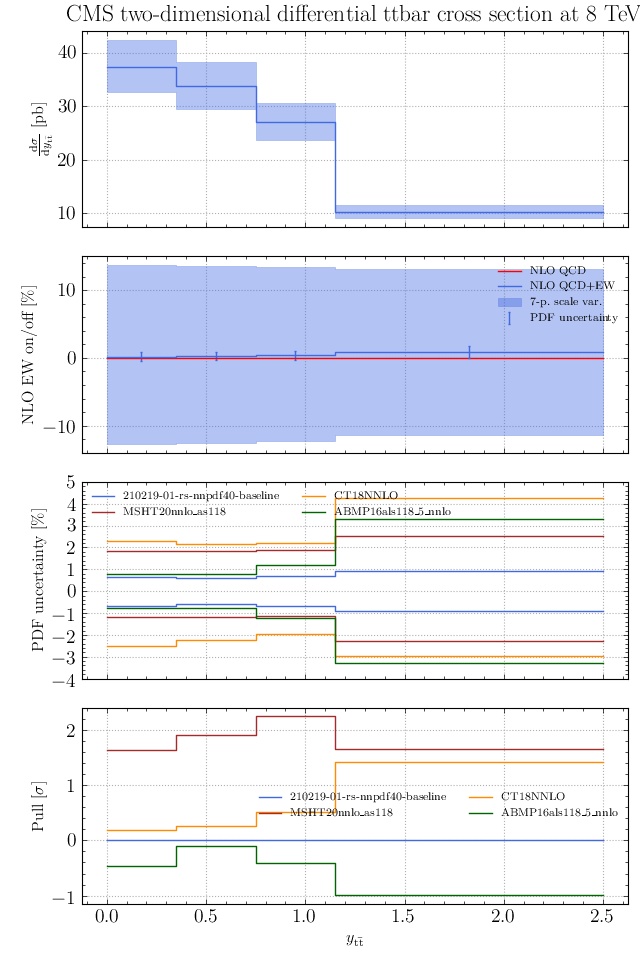
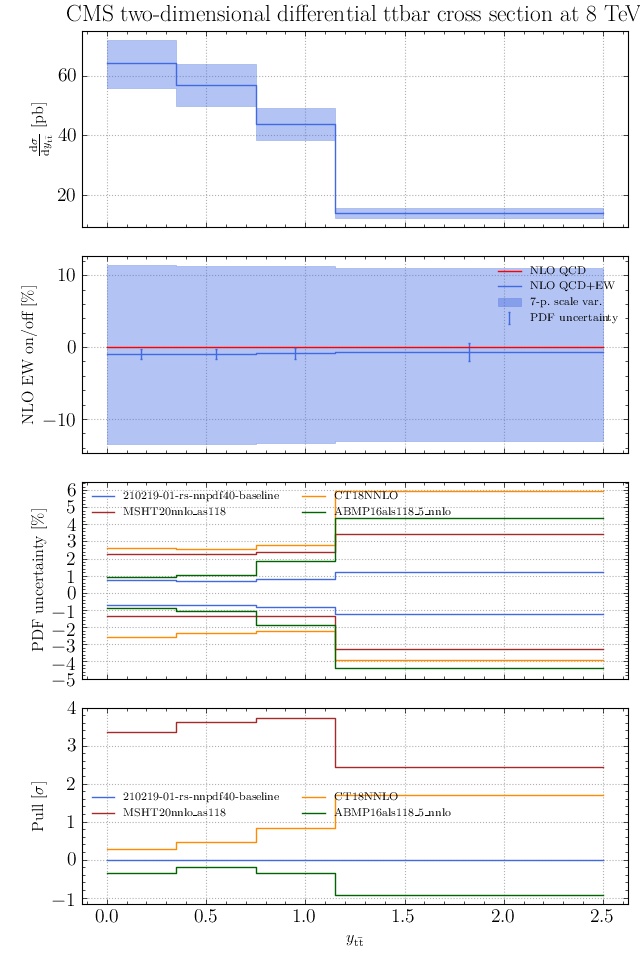
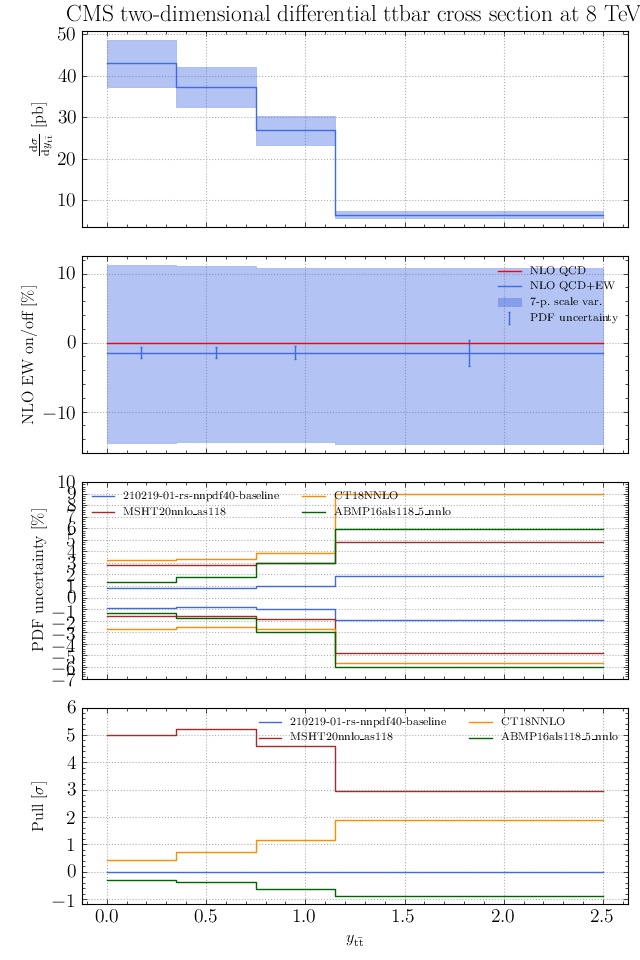
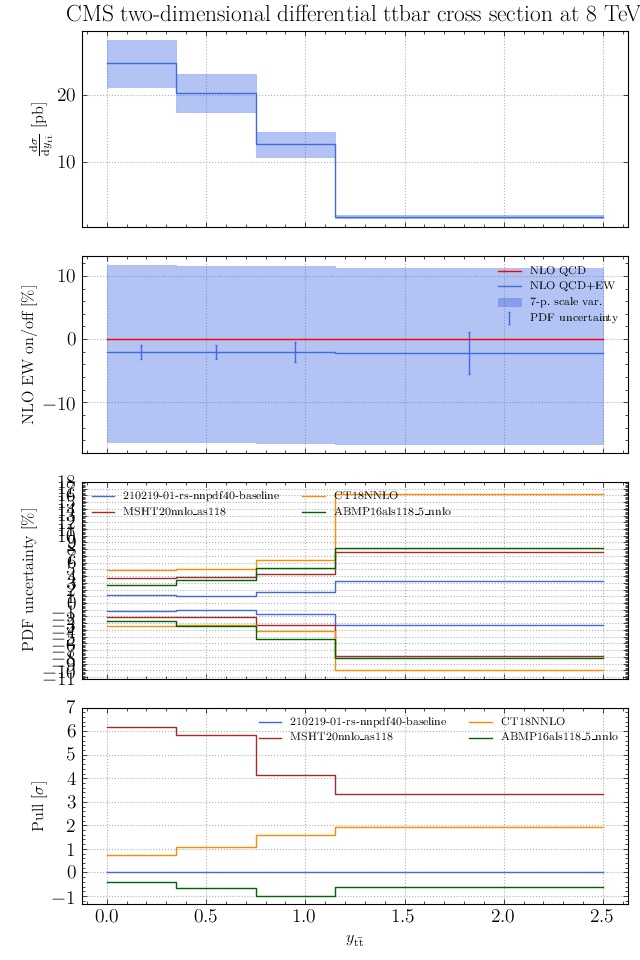
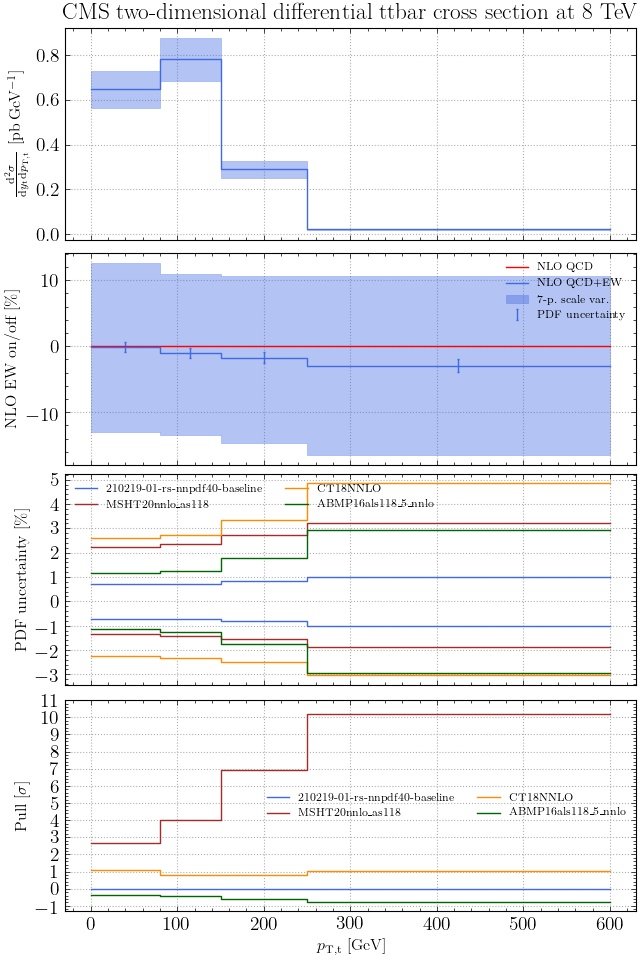
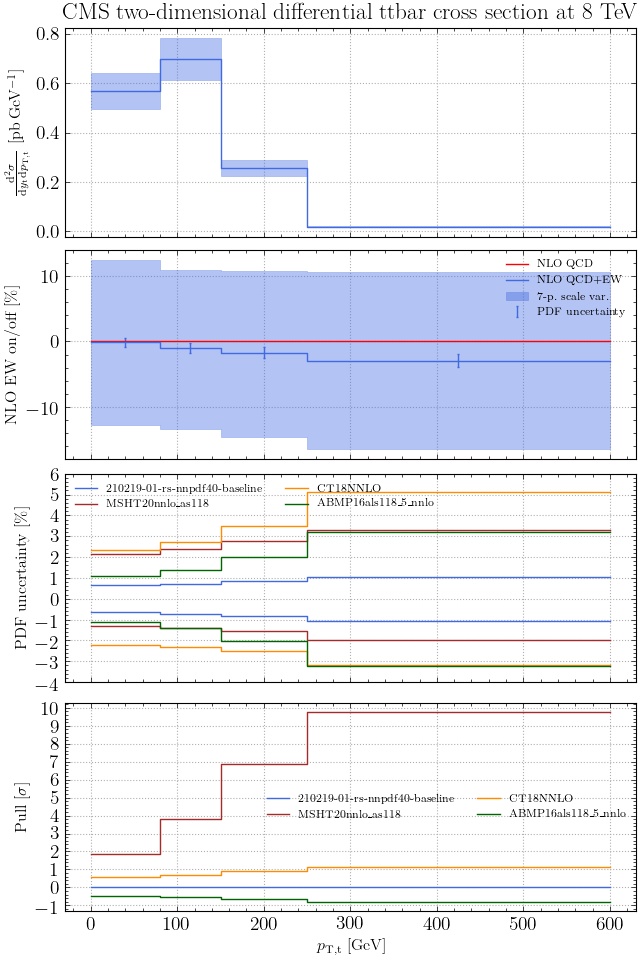
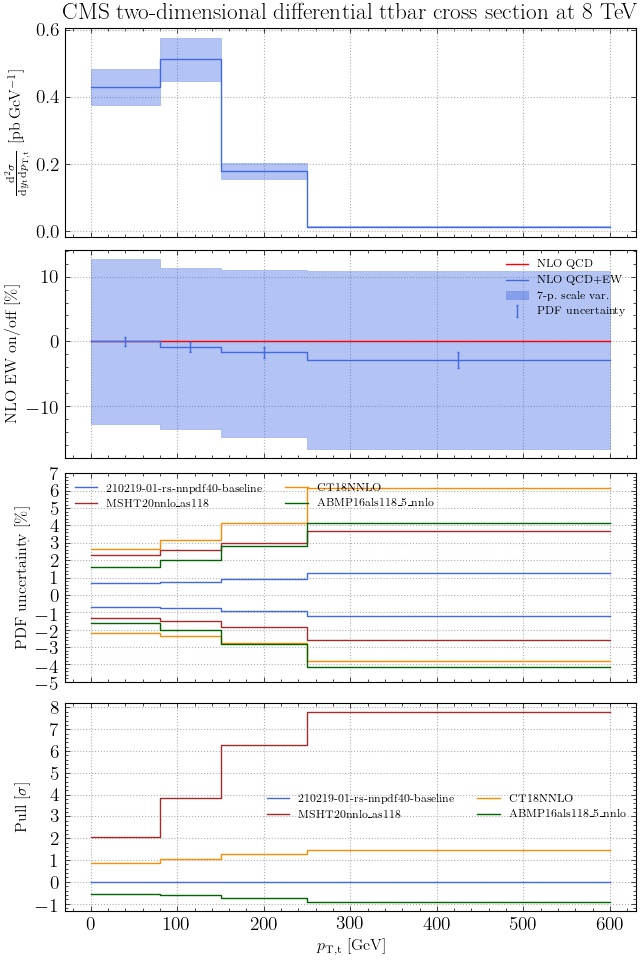
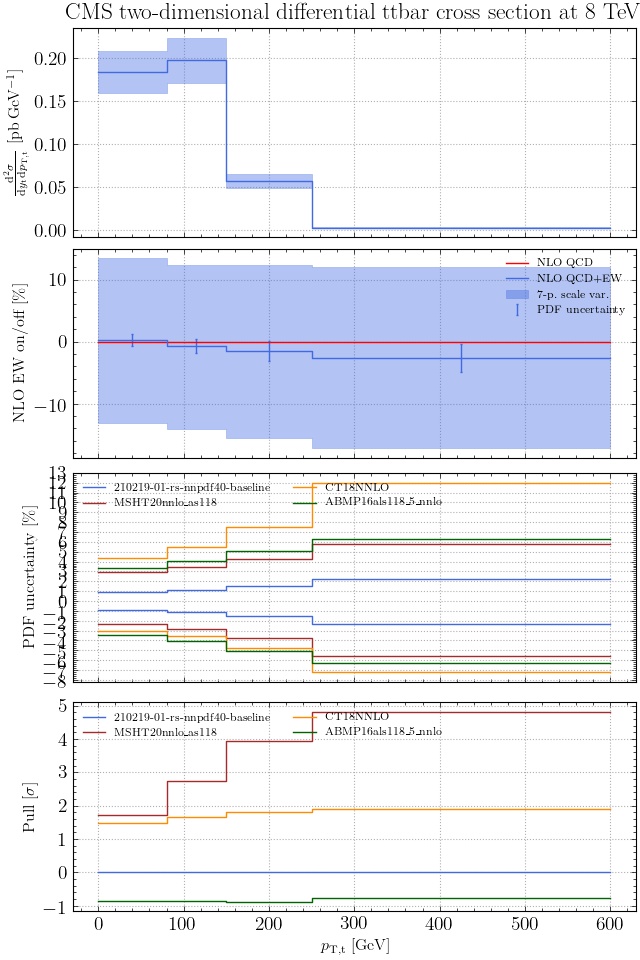
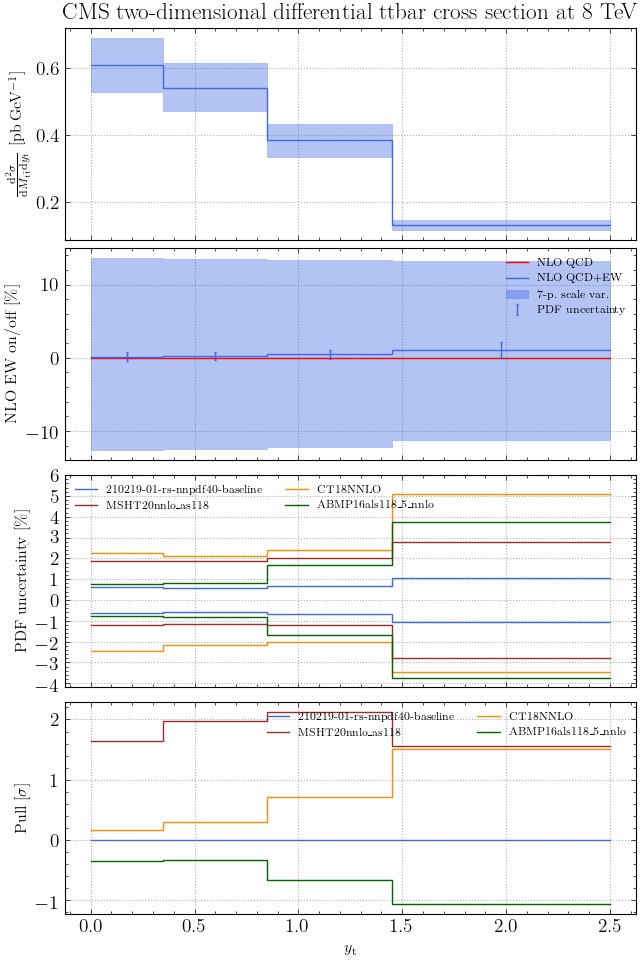
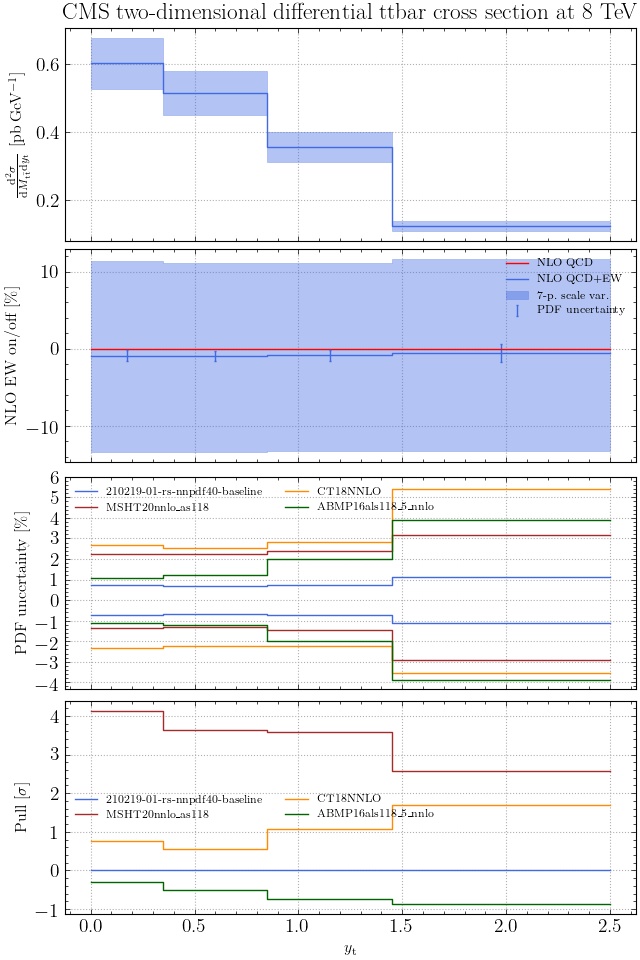
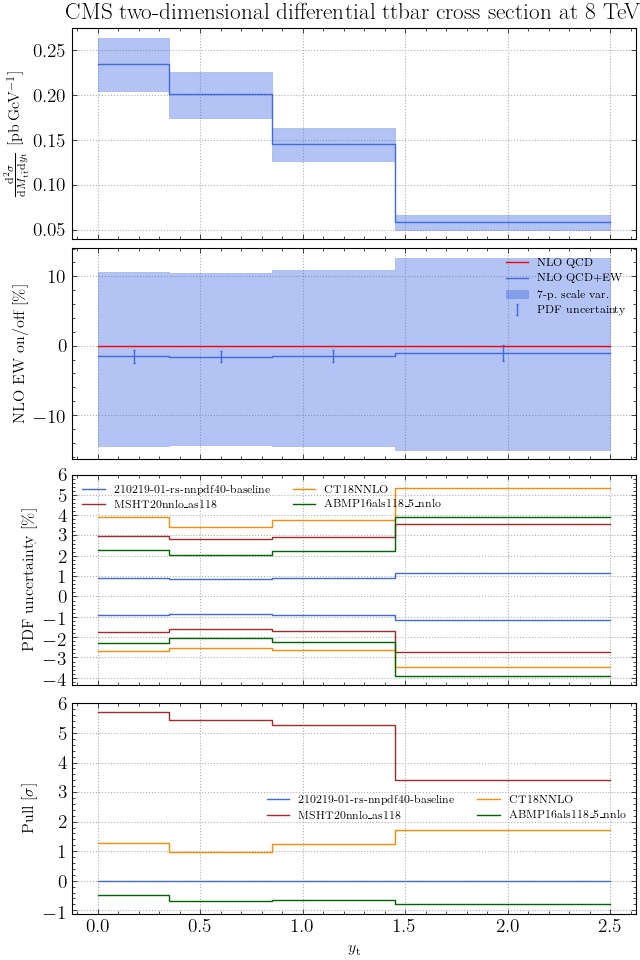
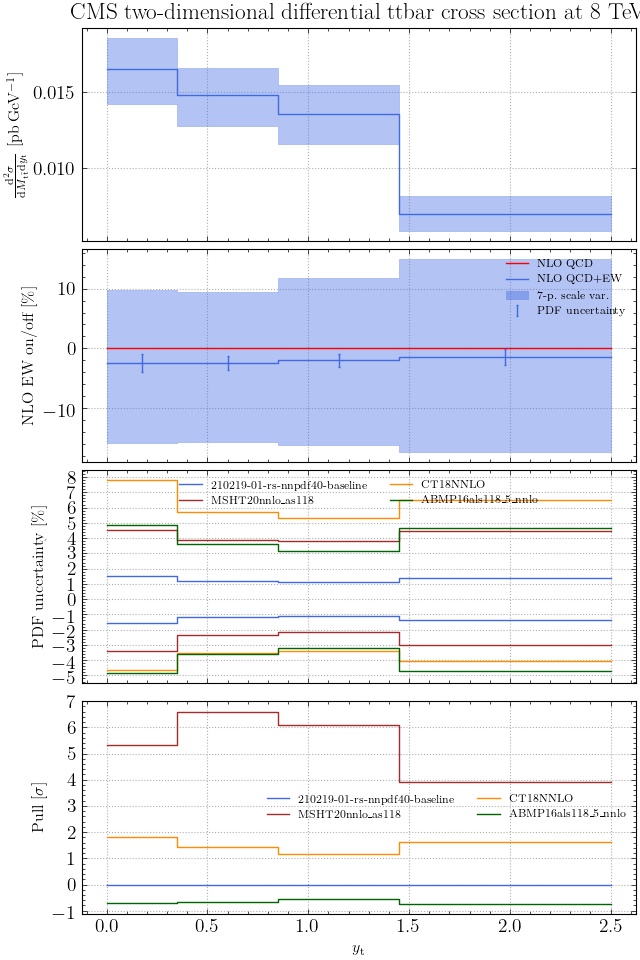
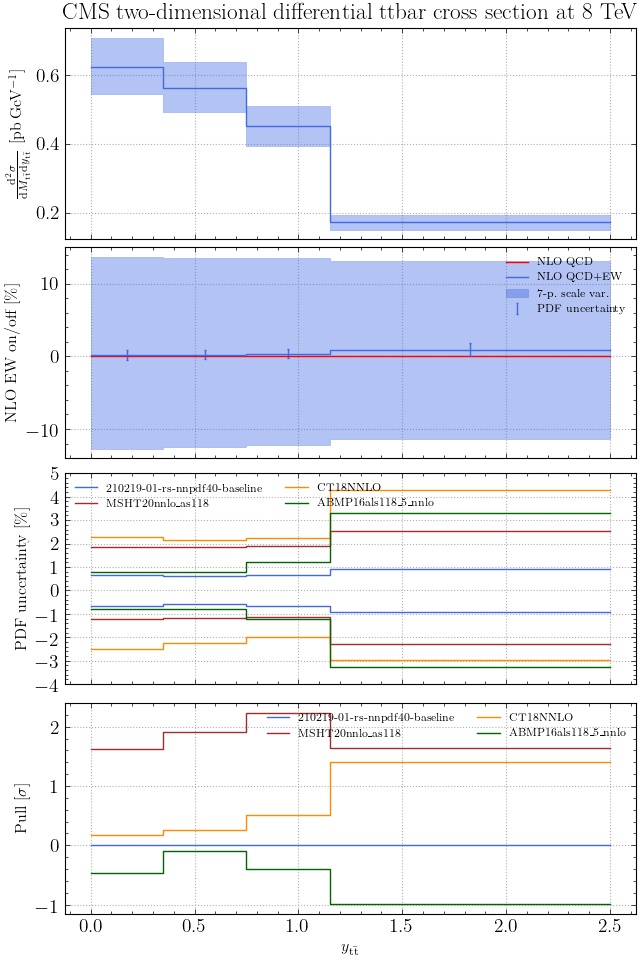
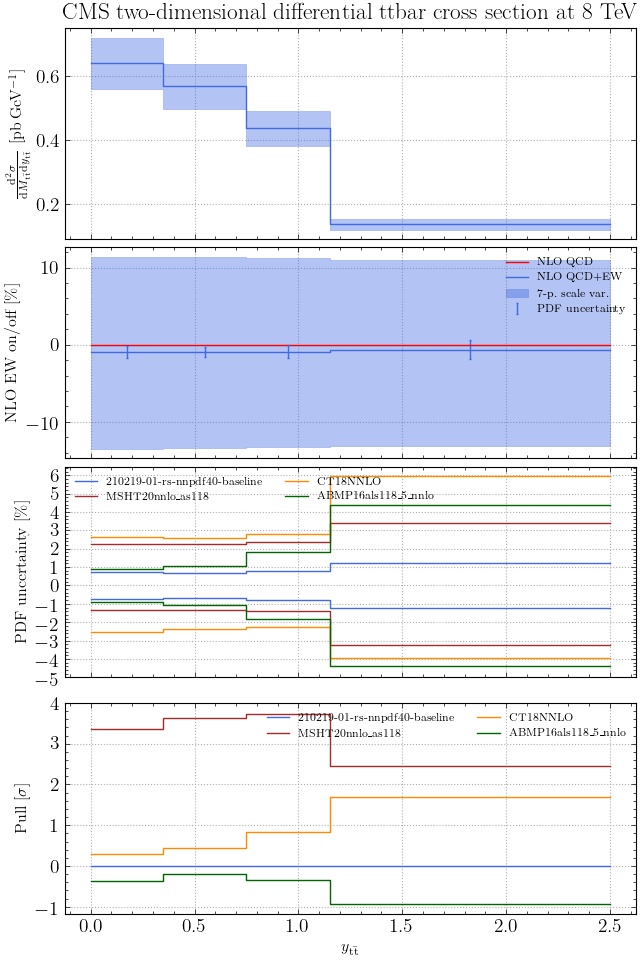
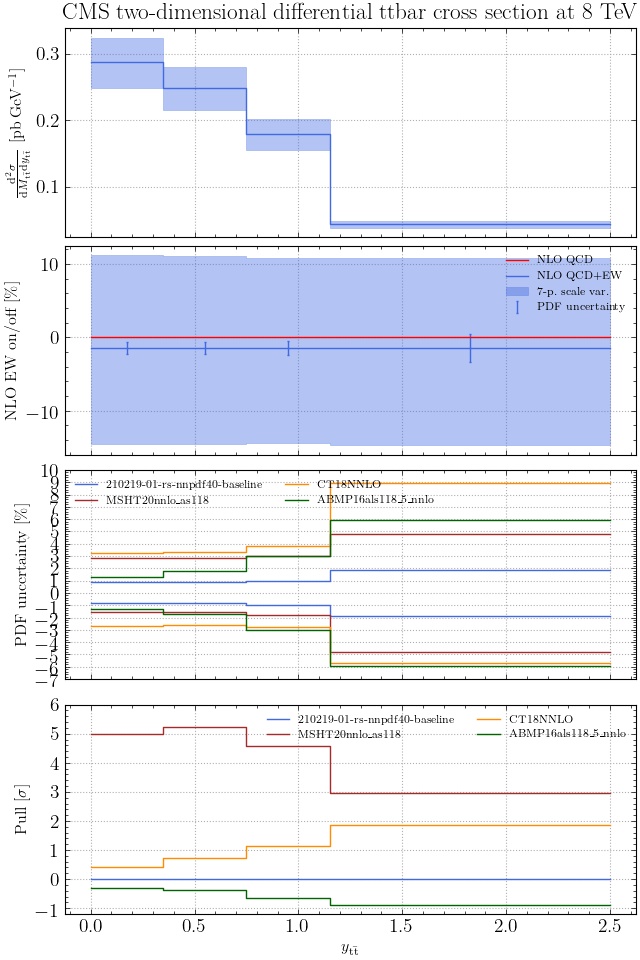
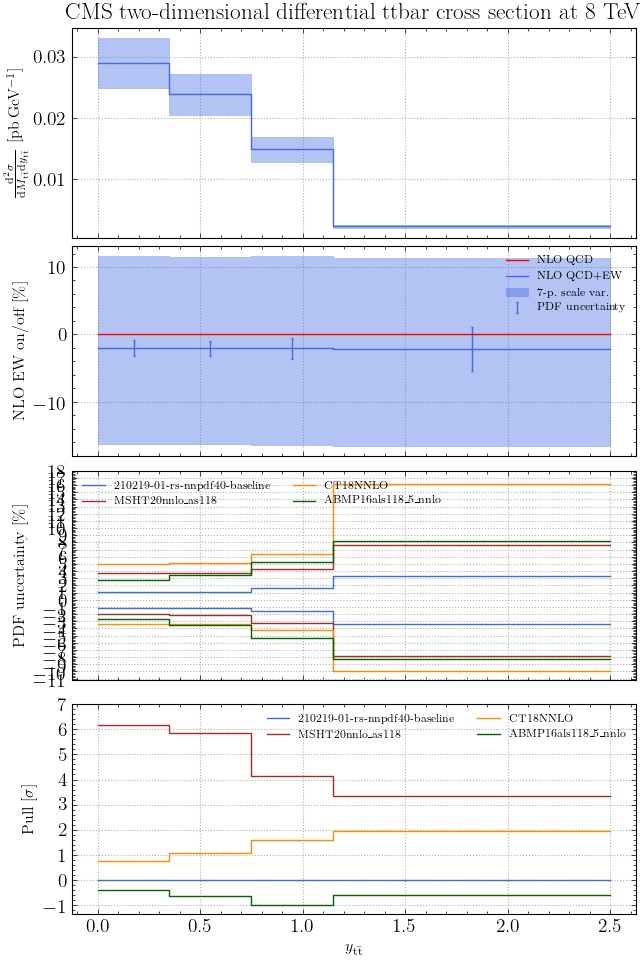
Result for the
TTM_TRAPgrid: