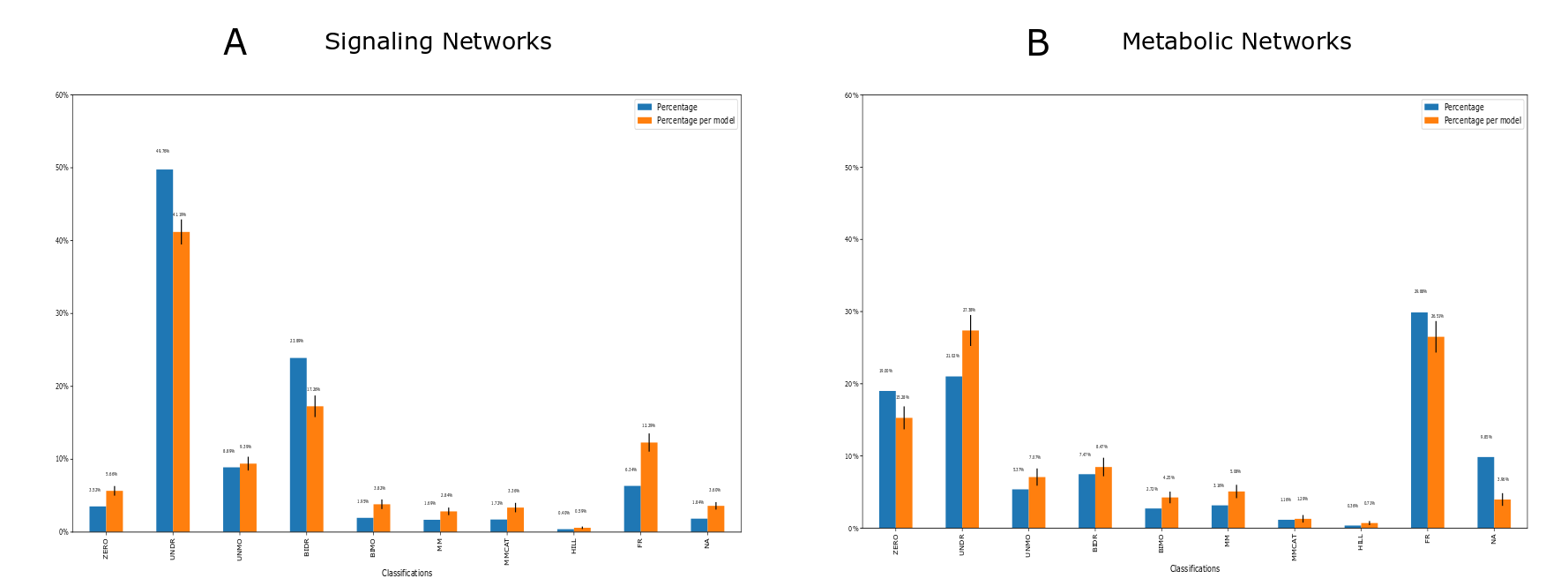
SBMLKinetics is a Python package to evaluate and classify kinetics in SBML models. There are many possible kinetics like the zeroth order, mass action, Michaelis-Menten, Hill kinetics and others. This work characterizes the kinetics in the BioModels Database as an example to improve modeling best practices. Our tool can analyze any data sets with SBML files as input. Users can also use this tool to compare different data sets. For instance, we compare the distribution of kinetics for the signaling and metabolic networks and find the substantial differences between two types of networks.
If you are using any of the code, please cite the article (https://bmcbioinformatics.biomedcentral.com/articles/10.1186/s12859-023-05380-3) and the PYPI web page (https://pypi.org/project/SBMLKinetics/).
pip install SBMLKinetics
Here is a classification example generated by SBMLKinetics:
Please see more examples in the documentation.
Please see the documentation at https://sunnyxu.github.io/SBMLKinetics/ for details.
- Install spyder3
- Clone the
SBMLKineticsrepository usinggit clone https://github.com/SunnyXu/SBMLKinetics - Create a virtual environment for the project.
cd SBMLKineticspython -m venv kvsource kv/Scripts/activate(Use "\" in windows.)pip install -r requirements.txtdeactivate
To verify the setup:
- Return to the
SBMLKineticsdirectory. source kv/Scripts/activate(Use "\" in windows.)export PYTHONPATH=`pwd`python tests/test_simple_sbml.py. The tests should run without error. (Use "\" in windows.)
cd SBMLKineticssource kv/bin/activate(Use "\" in windows.) When you're done, usedeactivate.
examples/tutorial.pyhas code illustrating usageSBMLKinetics/common/*.pyhas codes for the SmpleSBML (simple_sbml.py), Reaction (reaction.py), and KineticLaw (kinetic_law.py).