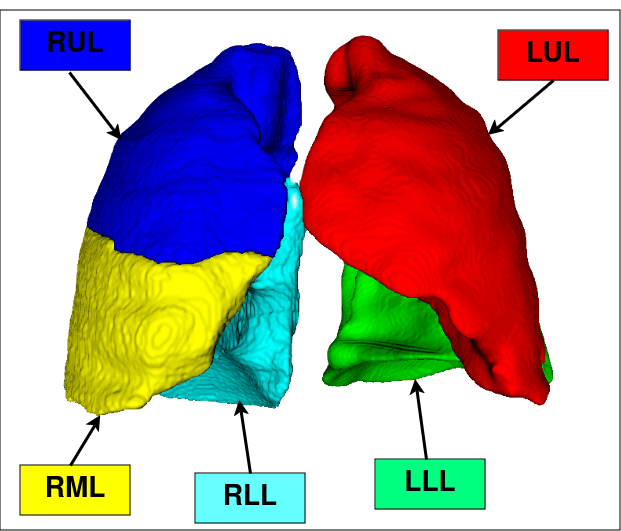
The development of efficient and robust algorithms for lung and lobe segmentation is essential for diagnosing and monitoring pulmonary diseases. However, obtaining manual or automatic annotations of lung lobes is challenging, especially in patients with severe abnormalities due to difficulty visualizing lobar fissures. This work aims to provide an automated lung lobe segmentation method using deep neural networks and probabilistic models, called LobePrior. Segmentation is performed in three stages: a coarse stage that processes images with reduced resolution; a high-resolution stage, in which specialized AttUNets compete for the segmentation of each lung lobe; and a final stage where post-processing is applied to the segmented lobes. Probabilistic models, constructed from label fusion, are responsible to carry anatomical information and guide the model in regions where severe abnormalities have caused segmentation failures. For additional resilience against abnormality presence, synthetic lesion generation was used as augmentation during training. The performance of the proposed approach was evaluated on LOLA11 (Grand Challenge) and three datasets with manual lobe annotations, in the presence of cancerous nodules and COVID-19 consolidations. Qualitative and quantitative results demonstrate that LobePrior achieved more accurate segmentations compared to manual ground truth, achieving state-of-the-art performance in the presence of severe abnormalities.
This tool was tested on Ubuntu 20.04. The following instructions refer to quickly running the tool by installing it with Miniconda and pip. The minimum recommended GPU memory is 12 GB for inference and 24 GB for training. The required dependencies are installed during setup and are listed in the requirements.txt file.
It is recommended to create a dedicated environment to run the LobePrior method to avoid interference with your current environment. To install Miniconda on Linux, follow the instructions available at: Miniconda.
To create a Miniconda environment named lobeprior, run:
conda create -n lobeprior python=3.9
To activate the lobeprior environment and install the required packages:
conda activate lobeprior
First, clone the repository and enter the LobePrior directory:
cd LobePrior
pip install .
Go to the Final folder setup section
Due to the large size of network weights, you need to go into the Releases in this repository, download the data.zip file, and put it inside the lobeprior folder. To To download the files, run:
wget https://github.com/MICLab-Unicamp/LobePrior/releases/download/LobePrior/data.zip
If this method of downloading the weights doesn't work for you, use this alternative link Data. The names of the folders and files must not be changed.
Extract the .ckpt files inside the LobePrior/weight folder and raw_images inside the LobePrior folder.
unzip data.zip
Install the dependencies
pip install -r requirements.txt
Finally, the directory containing LobePrior will look like this:
python predict.py -i input -o output
python predict.py -i input -o output -n
python predict_lung.py -i input -o output
python predict.py -i input -o output -p
To predict lung lobe segmentation using parallel computing, add -p and -nw to specify the number of processes
python predict.py -i input -o output -p -nw 4
If you wish to test the LobePrior method, you may use a public dataset referred to here as CoronaCases. The COVID-19 CT Lung and Infection Segmentation Dataset, available on Zenodo, provides 20 CT scans of COVID-19 patients. You can access and download the dataset directly via the following link:
LOCCA: Manual annotations on CT for lung LObes of COVID and CAncer patients or MICLab-Unicamp/LOCCA
- This work was accepted in CBEB 2024 (https://sbeb.org.br/cbeb2024).
-
Diagram of the method implemented using CCN and a priori information, incorporated into the network input as probabilistic models:
-
Segmentations using the LobePrior, nnUnet and LungMask methods on the CT image coronacases_007 (slice 150), from the Coronacases[1] dataset. Here, it is possible to visualize the segmentation errors (rectangular region) generated by each of the networks, in relation to the \textit{gold standard} (Fig. b):
-
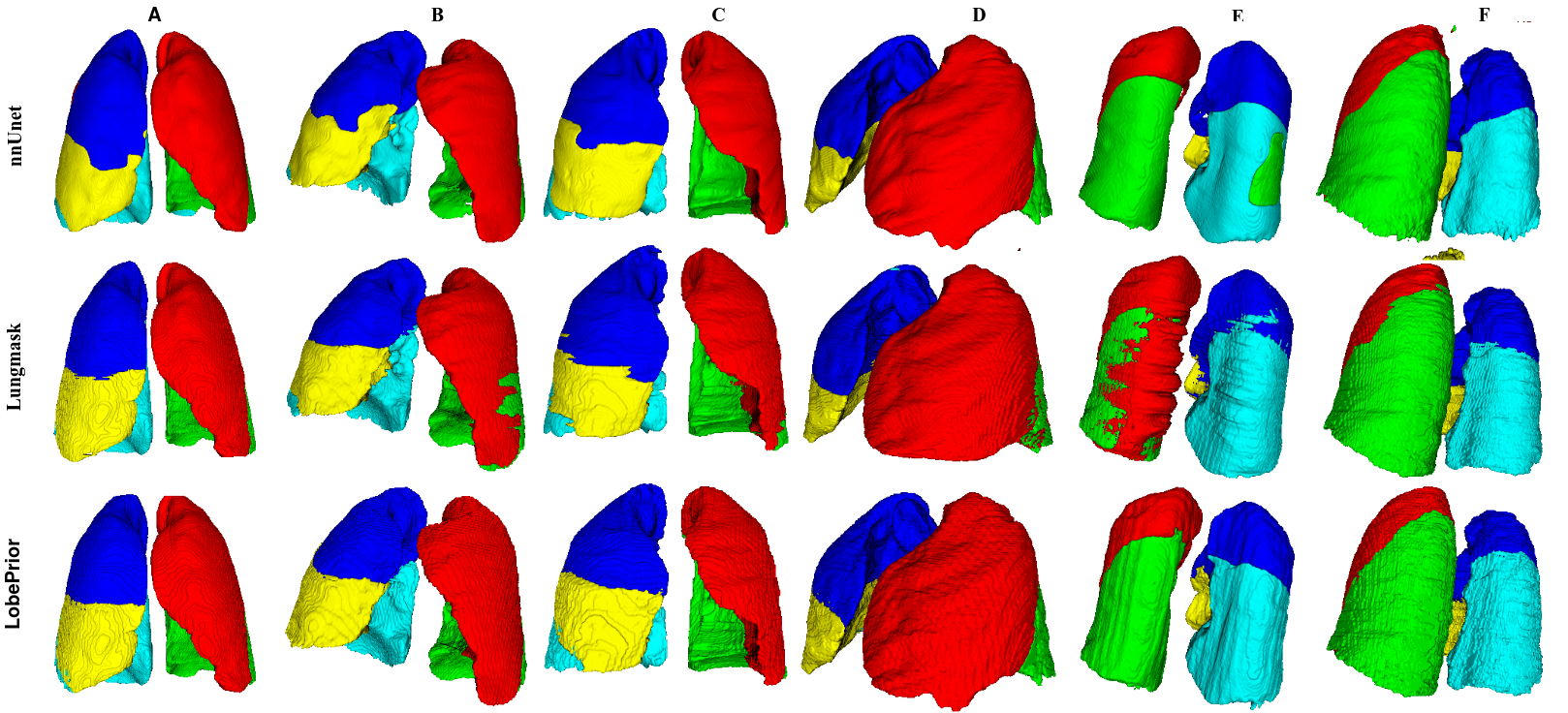
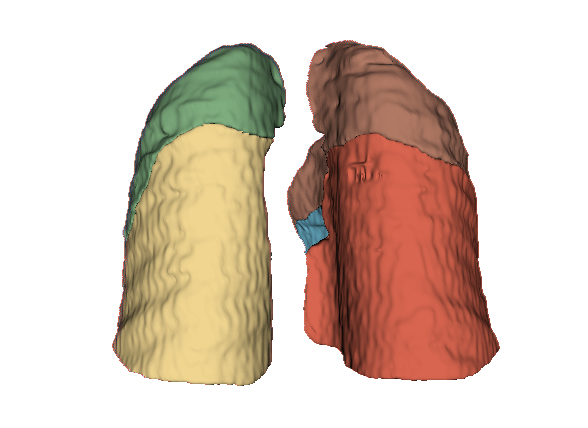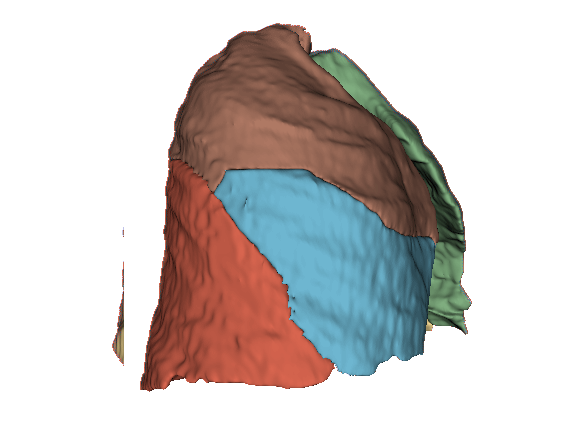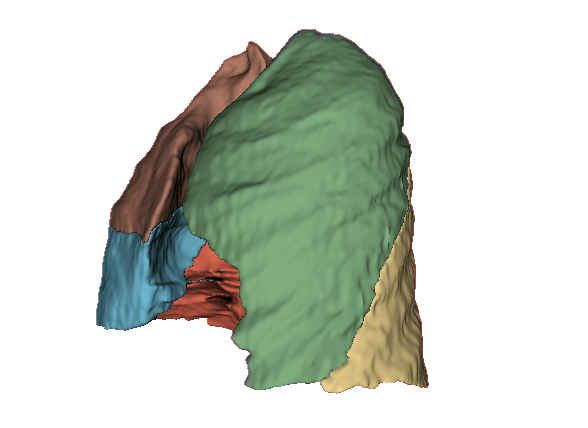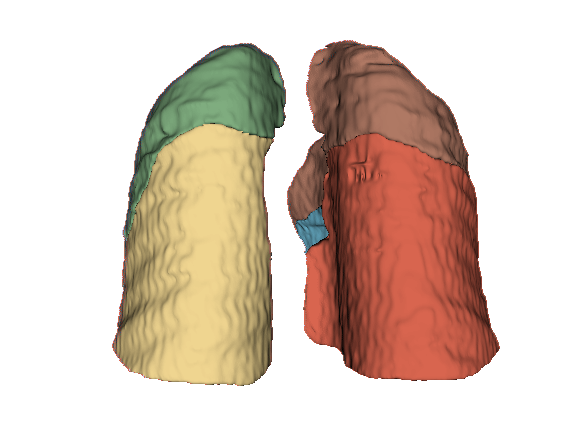
Qualitative evaluation using 3D representations in CT images of lungs from patients with severe injuries:
-
Qualitative evaluation using 3D representations in CT images of lungs from patients with severe injuries:
Deep learning with probabilistic models for segmenting lung lobes on computed tomography images with severe abnormalities
Initial project presented at the XXIX Congresso Brasileiro de Engenharia Biomédica (CBEB) 2024 (https://sbeb.org.br/cbeb2024). This manuscript represents an extended and enhanced version of the work previously published as a conference paper, with the following additional contributions:
-
Synthetic lesion insertion: a mechanism for simulating pulmonary lesions was incorporated to increase the model’s robustness when dealing with severe anatomical abnormalities;
-
Creation of unbiased probabilistic models: probabilistic maps of the lobar regions were developed based on normal cases, in a systematic manner and independently of the training data, thus avoiding bias and promoting a more generalizable anatomical representation;
-
Expanded dataset: expanded from 44 training and 14 testing samples to 150 training and 85 testing samples, now including cases with severe lung lesions;
-
Test set: added a new set including severe lesions to evaluate performance under challenging conditions;
-
Improved loss functions: changed from Focal Loss to Dice Loss for better optimization. The loss functions were refined to better guide the learning process, especially in regions with ambiguous or incomplete fissures;
-
More comprehensive evaluation: the method was tested on a larger number of scans, including cases with multiple and extensive lesions, enabling a more rigorous validation of the proposed approach;
-
Architecture redesign: replaced five independent AttUNets (without weight sharing and inefficient) with a single network comprising seven decoders with shared weights.
@ARTICLE{redu_ORXJKS_2025,
author = {Jean Antonio Ribeiro and Leticia Rittner and Diedre Santos do Carmo and Simone Appenzeller and Ricardo Siufi Magalhães and Sergio San Juan Dertkigil and Fabiano Reis},
journal={IEEE Data Descriptions},
title={Descriptor: Manually Annotated CT Dataset of Lung Lobes in COVID-19 and Cancer Patients (LOCCA)},
year = {2025},
pages = {1-8},
doi={10.1109/IEEEDATA.2025.3577999}
}
@ARTICLE{RibeiroLOCCA2025,
author={Ribeiro, Jean A. and Carmo, Diedre S. Do and Reis, Fabiano and Magalhães, Ricardo S. and Dertkigil, Sergio S. J. and Appenzeller, Simone and Rittner, Leticia},
journal={IEEE Data Descriptions},
title={Descriptor: Manually Annotated CT Dataset of Lung Lobes in COVID-19 and Cancer Patients (LOCCA)},
year={2025},
volume={2},
number={},
pages={239-246},
keywords={Lungs;Computed tomography;Annotations;Lung cancer;Biomedical imaging;Lesions;Image segmentation;Manuals;COVID-19;Three-dimensional displays;Cancer;computed tomography (CT) images;COVID-19;dataset;manual annotation for lung lobes},
doi={10.1109/IEEEDATA.2025.3577999}
}
@ARTICLE{CBEB2024,
title = {Deep learning with probabilistic models for segmenting lung lobes on computed tomography images with severe abnormalities},
author = {Jean Antonio Ribeiro and Diedre Santos do Carmo and Fabiano Reis and Leticia Rittner},
journal = {CBEB 2024},
pages = {1-6},
year = {2024},
}
@ARTICLE{review2022,
title = {{A Systematic Review of Automated Segmentation Methods and Public Datasets for the Lung and its Lobes and Findings on Computed Tomography Images}},
author={Diedre Santos do Carmo and Jean Antonio Ribeiro and Sergio Dertkigil and Simone Appenzeller and Roberto Lotufo and Leticia Rittner},
journal={Yearbook of Medical Informatics},
volume={31},
number={01},
pages={277-295},
year={2022},
doi = {10.1055/s-0042-1742517}
}