-
Notifications
You must be signed in to change notification settings - Fork 1
Description
Pointers to useful references in gene regulation
-
nucleosome remodellers,AR 2009
Therefore, chromatin structure not only provides a packaging solution, but also an opportunity for regulation
-
Topological associating domains (TADs), Science Advances 2018
-
ncRNA are promiscuous regulators who dip its hands in all aspects: 1. Chromatin remodeling. 2. Regulation of transcription 3. Regulation of transcript stability and translation. In other words, a unifying framework is absent.
-
Integrative analyses of 111 human epigenomes, nature2015, citing chromHMM
Videos:
paradigm around RNAPII:
- Basal transcription is present around TATA-based promoters, which recruits the formation of per-initiation complex
- Transcription is further promoted in the present of mediator complex
- trans-cis interplay is complex and involves things like p300/PRC/mediator/SWI/SNF.
- most of these are qualitative arguments based on Western Blots/Southern Blots/Biochemistry/Molecular-Cloning/Structural Analyses and lacking a quantitative basis.
MISC
databases:
-
ENCODE: Encyclopedia of DNA elements
-
Roadmap epigenetics
-
Blueprint
-
PCSD: Plant Chromatin State Database
-
Semi-Automated Genome Annotation (SAGA) softwares: chromHMM (762 cite for its 2012 original correspondence, see WoK citation report), Segway (DBN-based, 1 cite bioRxiv paper) . Neither outlines the inference framework and are mostly details of application.
Experiments
- First ChIP-Seq, 43 pages of supplements
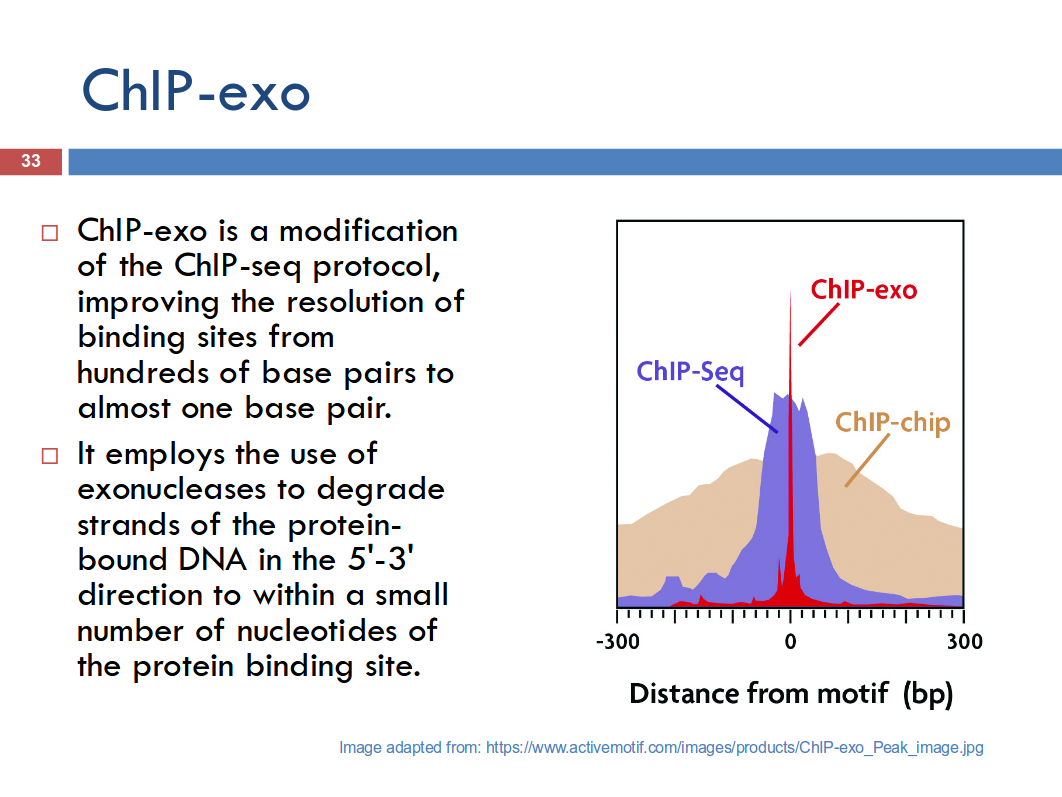
(from lec slides) (http://www.activemotif.com/catalog/704/epigenetic-services-1)