-
Notifications
You must be signed in to change notification settings - Fork 0
Description
Hi,
Thanks for creating this wrapper!
I'm running PEER using MOFA framework and comparing the results against the original PEER package. However, I do find discrepancies in the results. I assumed that the raw factors could be different between approaches but had similar effects on the eQTL analysis downstream. That was not the case.
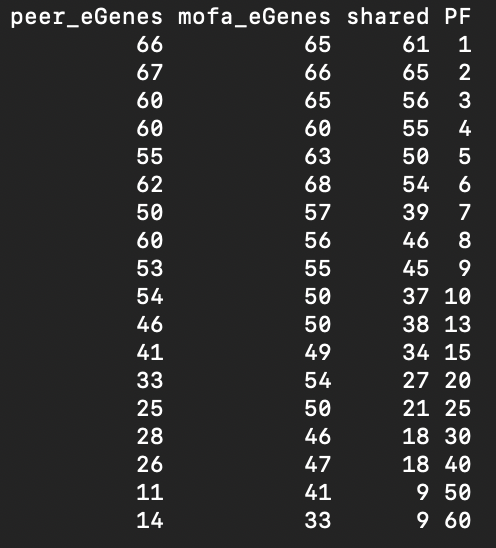
In the table, I'm showing the number of eGenes mapped for the same specific region with the two approaches. With the first factors (around 1-5) the eGenes found were very similar, but with greater factors, the results seem very different.
Is there any theoretical reason why the results are not consistent between approaches? I thought that with one view MOFA was effectively PEER and because of that gave identical results, but it is not the case with my data.
Thanks,
Cristian